# test function factories
study %>%
tidyplot(treatment, score, color = treatment) %>%
add_error_bar() %>%
add_sd_bar() %>%
add_range_bar() %>%
add_ci95_bar() %>%
add_data_points()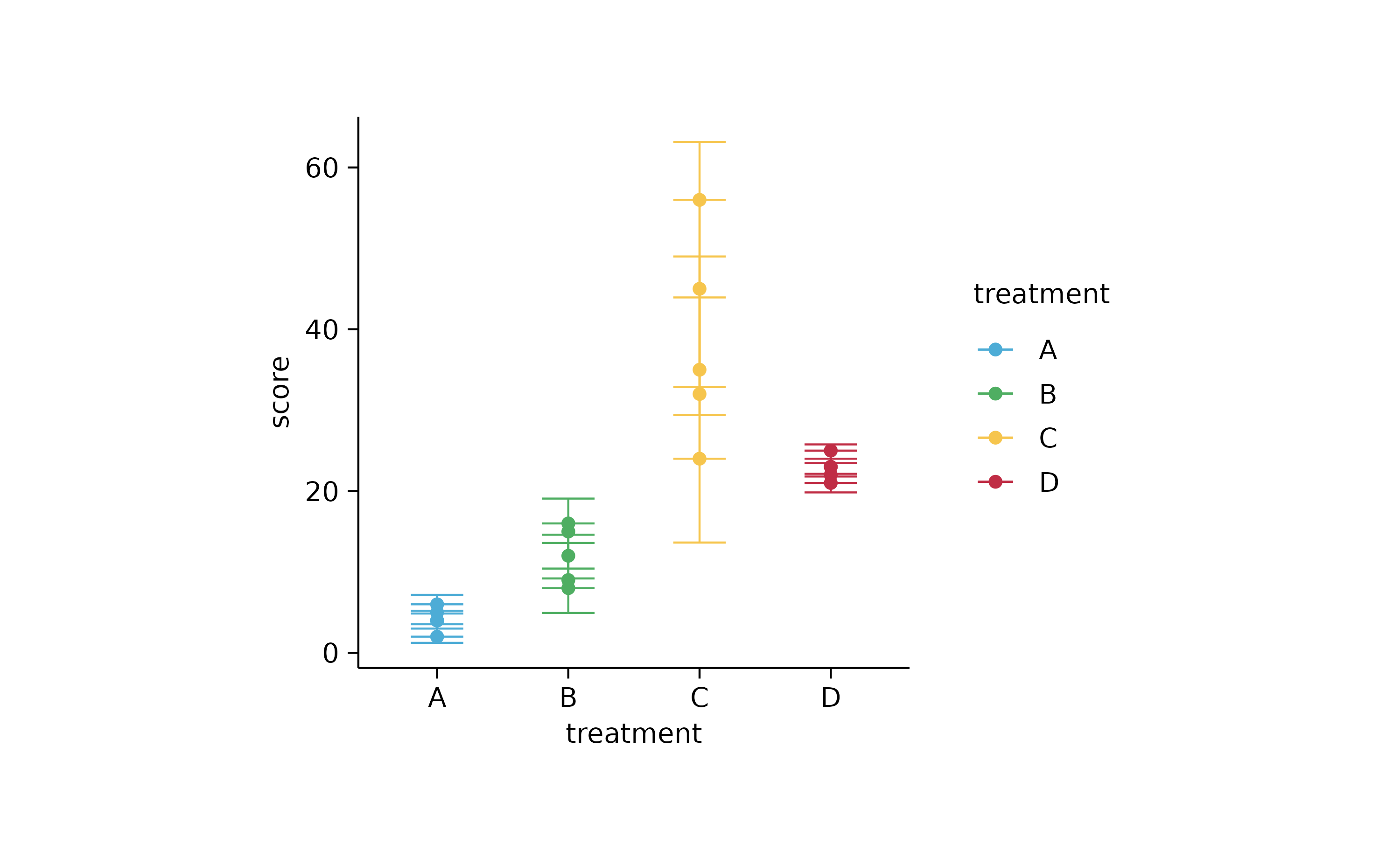
study %>%
tidyplot(score, treatment, color = treatment) %>%
add_error_bar() %>%
add_sd_bar() %>%
add_range_bar() %>%
add_ci95_bar() %>%
add_data_points()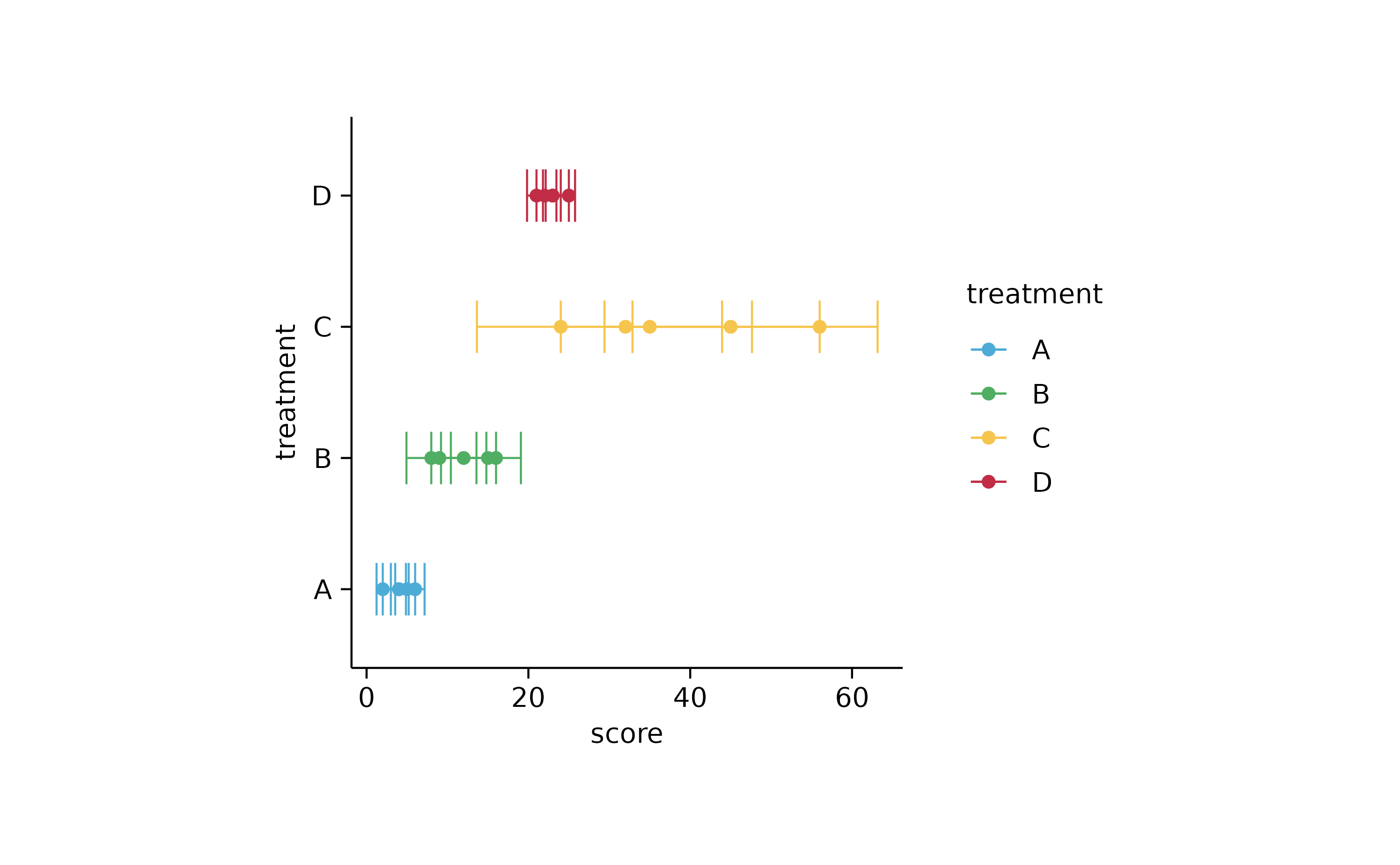
study %>%
tidyplot(treatment, score) %>%
add_error_ribbon() %>%
add_sd_ribbon() %>%
add_range_ribbon() %>%
add_ci95_ribbon() %>%
add_data_points()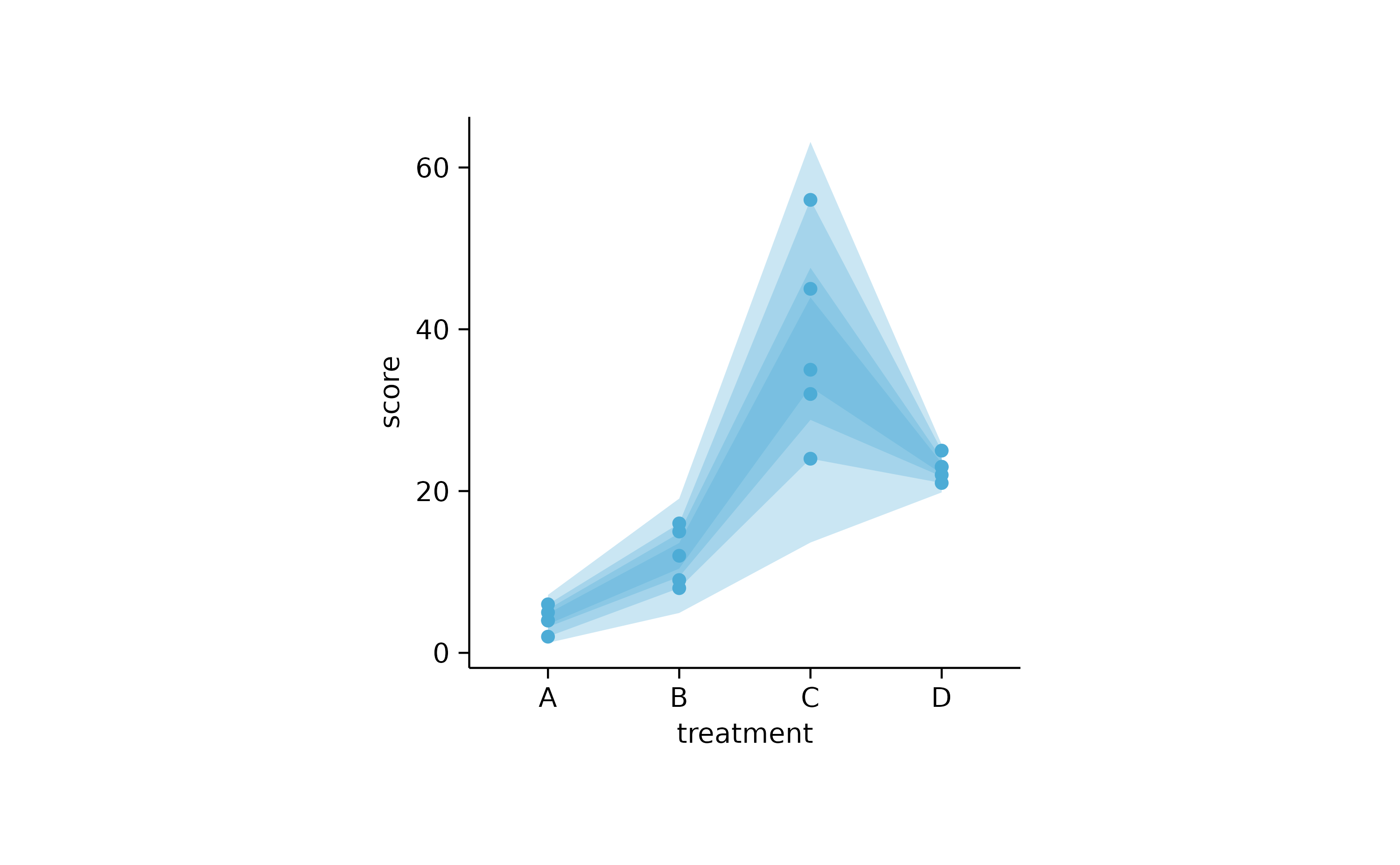
study %>%
tidyplot(score, treatment) %>%
add_error_ribbon() %>%
add_sd_ribbon() %>%
add_range_ribbon() %>%
add_ci95_ribbon() %>%
add_data_points()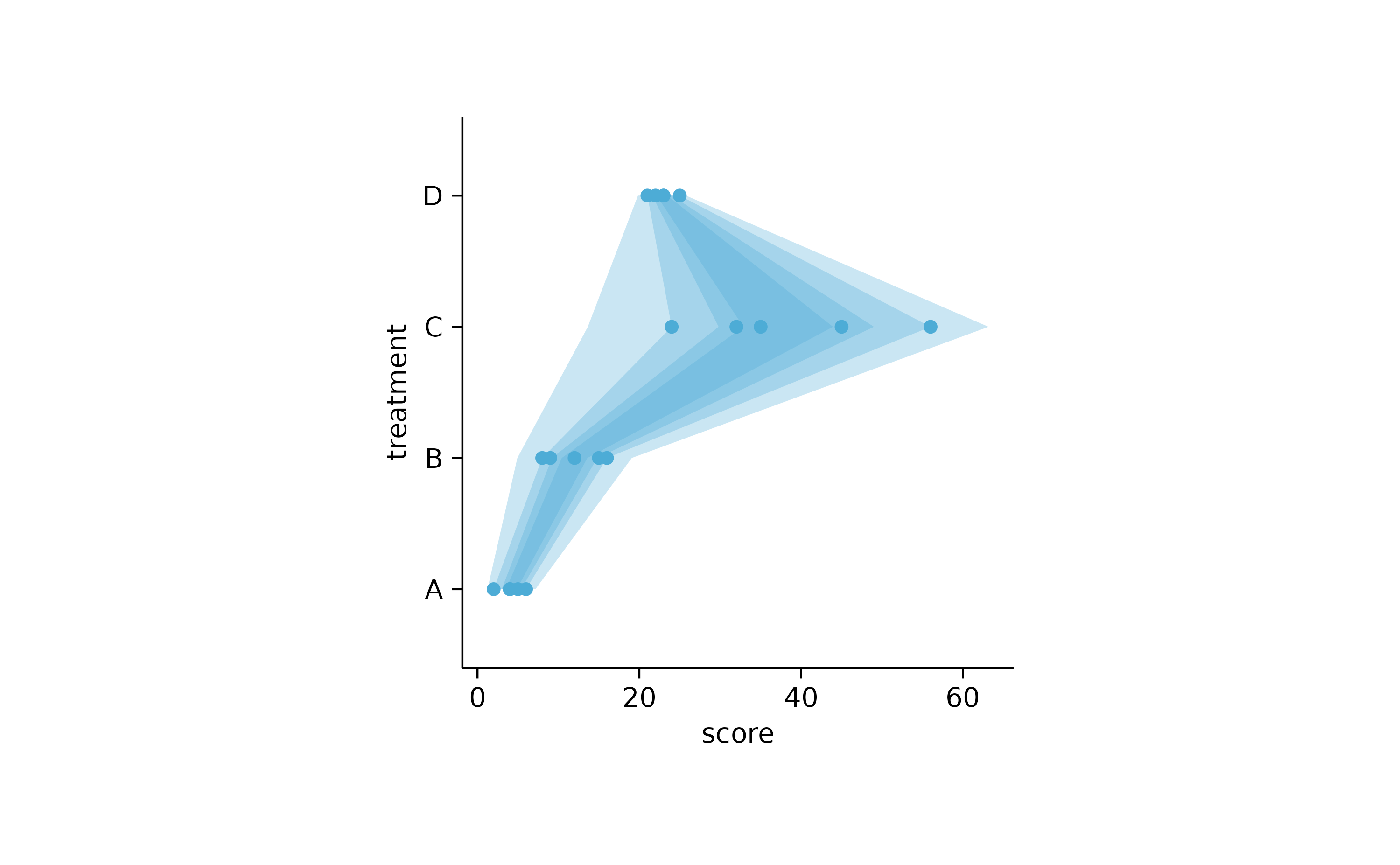
study %>%
tidyplot(treatment, score) %>%
add_mean_bar(alpha = 0.3) %>%
add_mean_dash() %>%
add_mean_dot() %>%
add_mean_value() %>%
add_mean_line() %>%
add_mean_area(alpha = 0.2, fill = "green") %>%
add_data_points()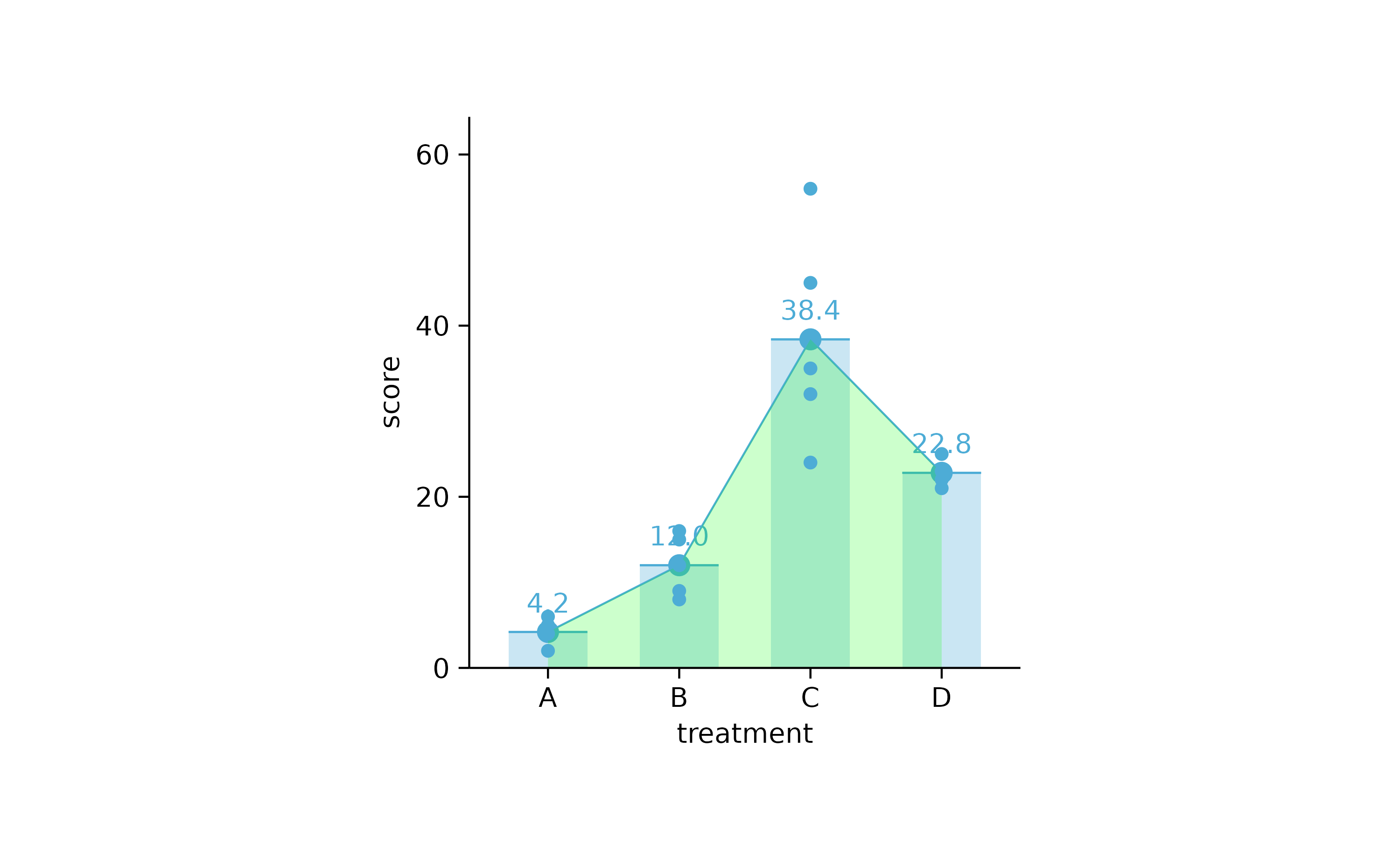
study %>%
tidyplot(score, treatment) %>%
add_mean_bar(alpha = 0.3) %>%
add_mean_dash() %>%
add_mean_dot() %>%
add_mean_value() %>%
add_mean_line() %>%
add_mean_area(alpha = 0.2, fill = "green") %>%
add_data_points()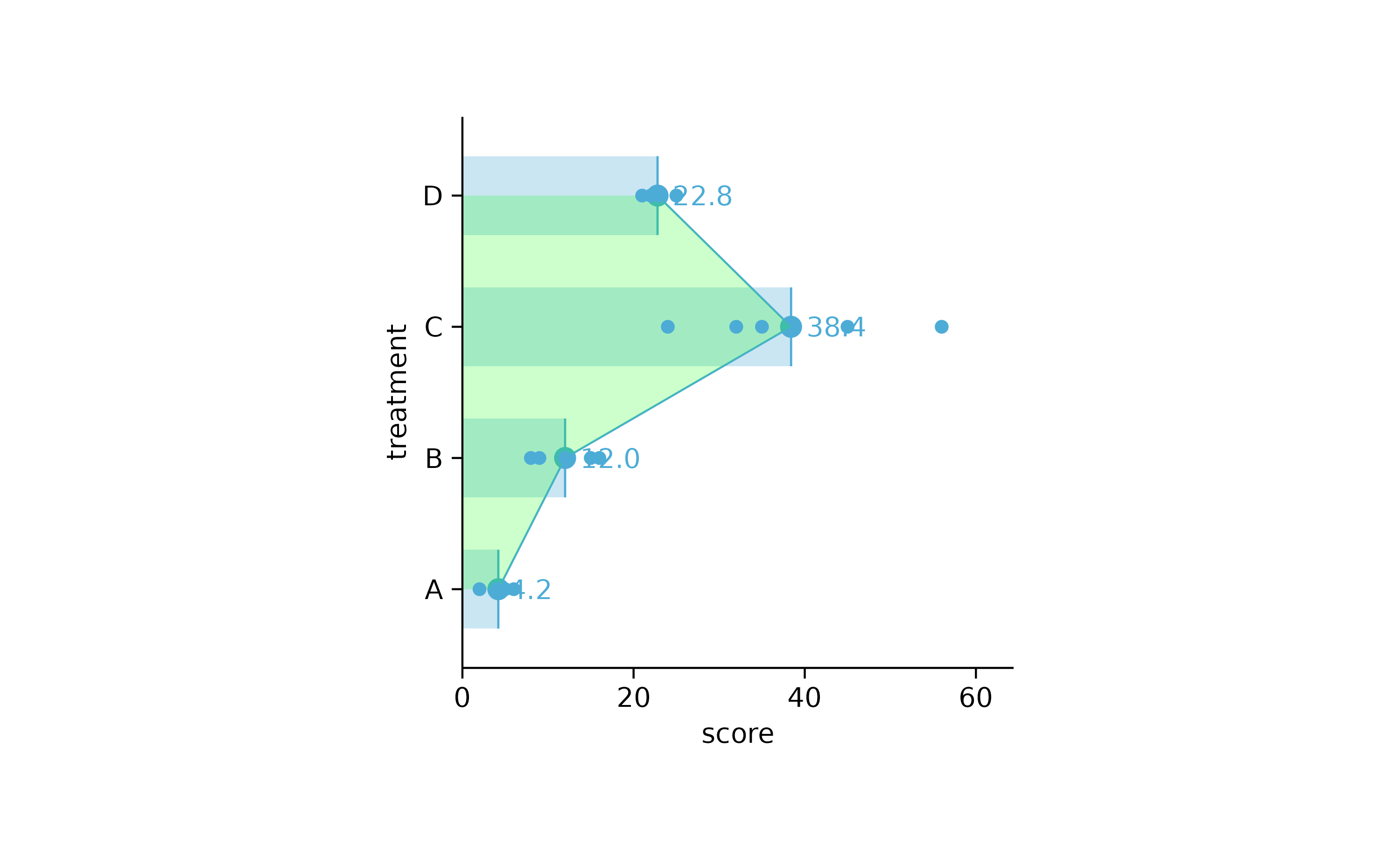
study %>%
tidyplot(treatment, score) %>%
add_median_bar(alpha = 0.3) %>%
add_median_dash() %>%
add_median_dot() %>%
add_median_value() %>%
add_median_line() %>%
add_median_area(alpha = 0.2, fill = "green") %>%
add_data_points()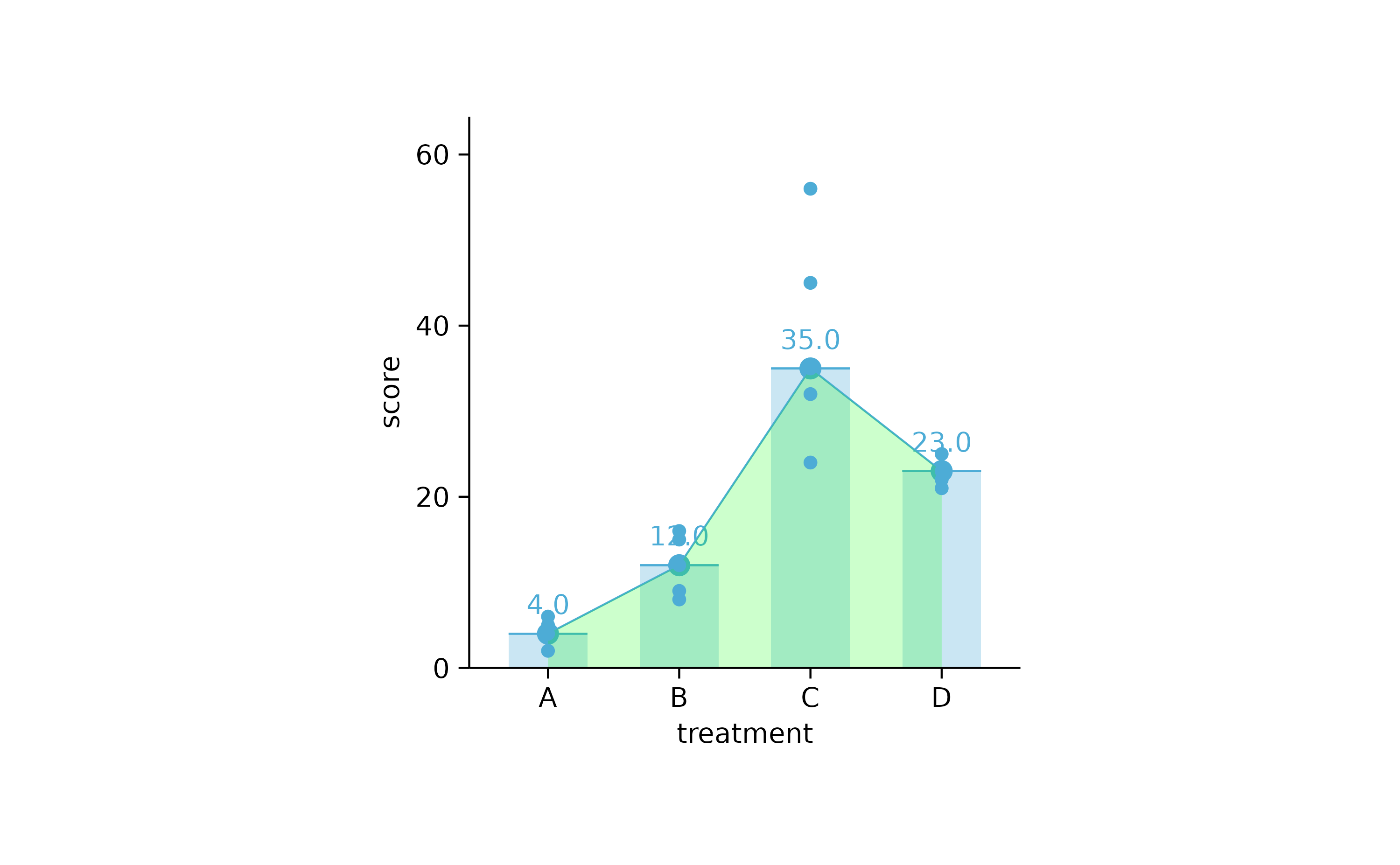
study %>%
tidyplot(score, treatment) %>%
add_median_bar(alpha = 0.3) %>%
add_median_dash() %>%
add_median_dot() %>%
add_median_value() %>%
add_median_line() %>%
add_median_area(alpha = 0.2, fill = "green") %>%
add_data_points()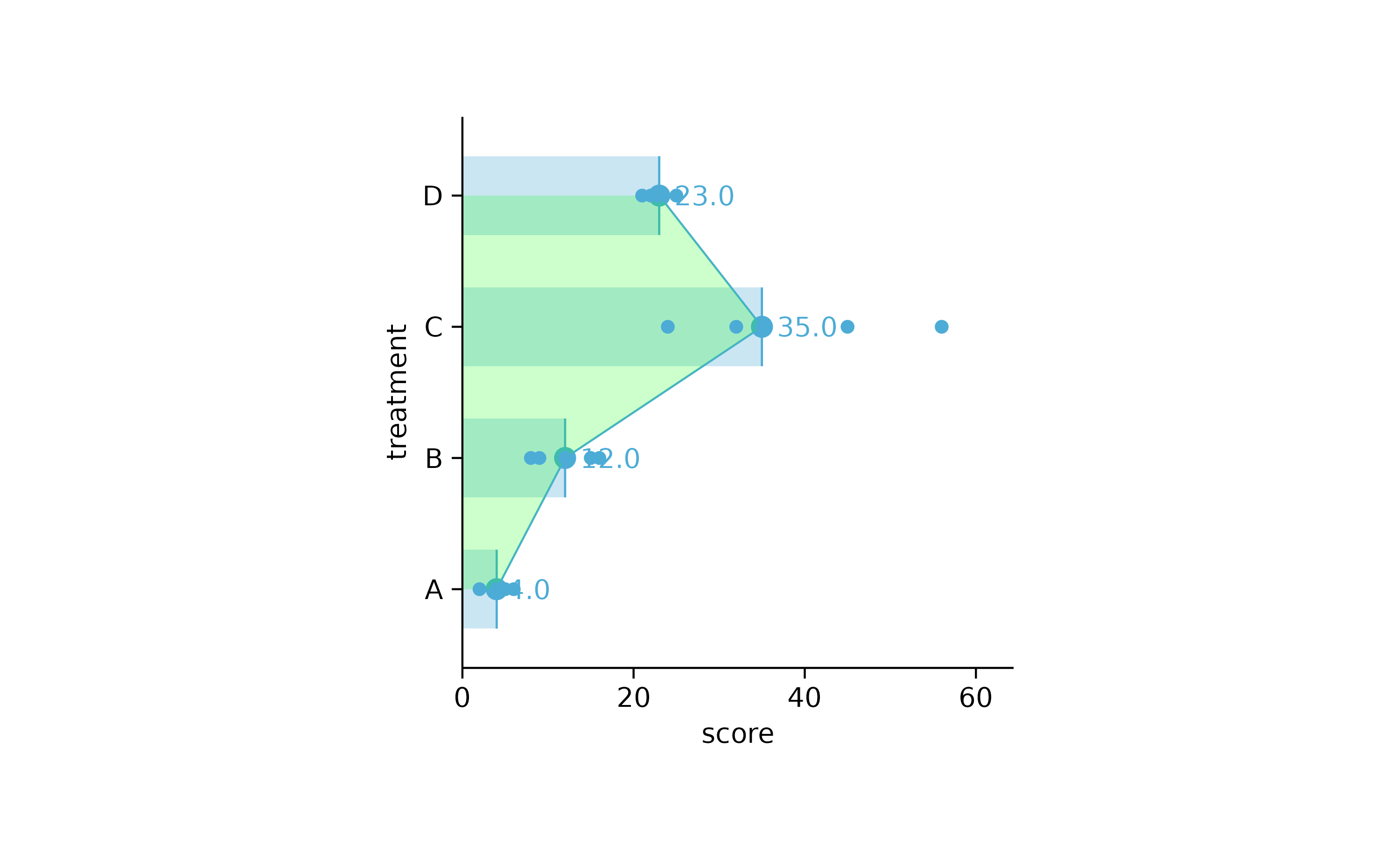
study %>%
tidyplot(treatment, score) %>%
add_sum_bar(alpha = 0.3) %>%
add_sum_dash() %>%
add_sum_dot() %>%
add_sum_value() %>%
add_sum_line() %>%
add_sum_area(alpha = 0.2, fill = "green") %>%
add_data_points()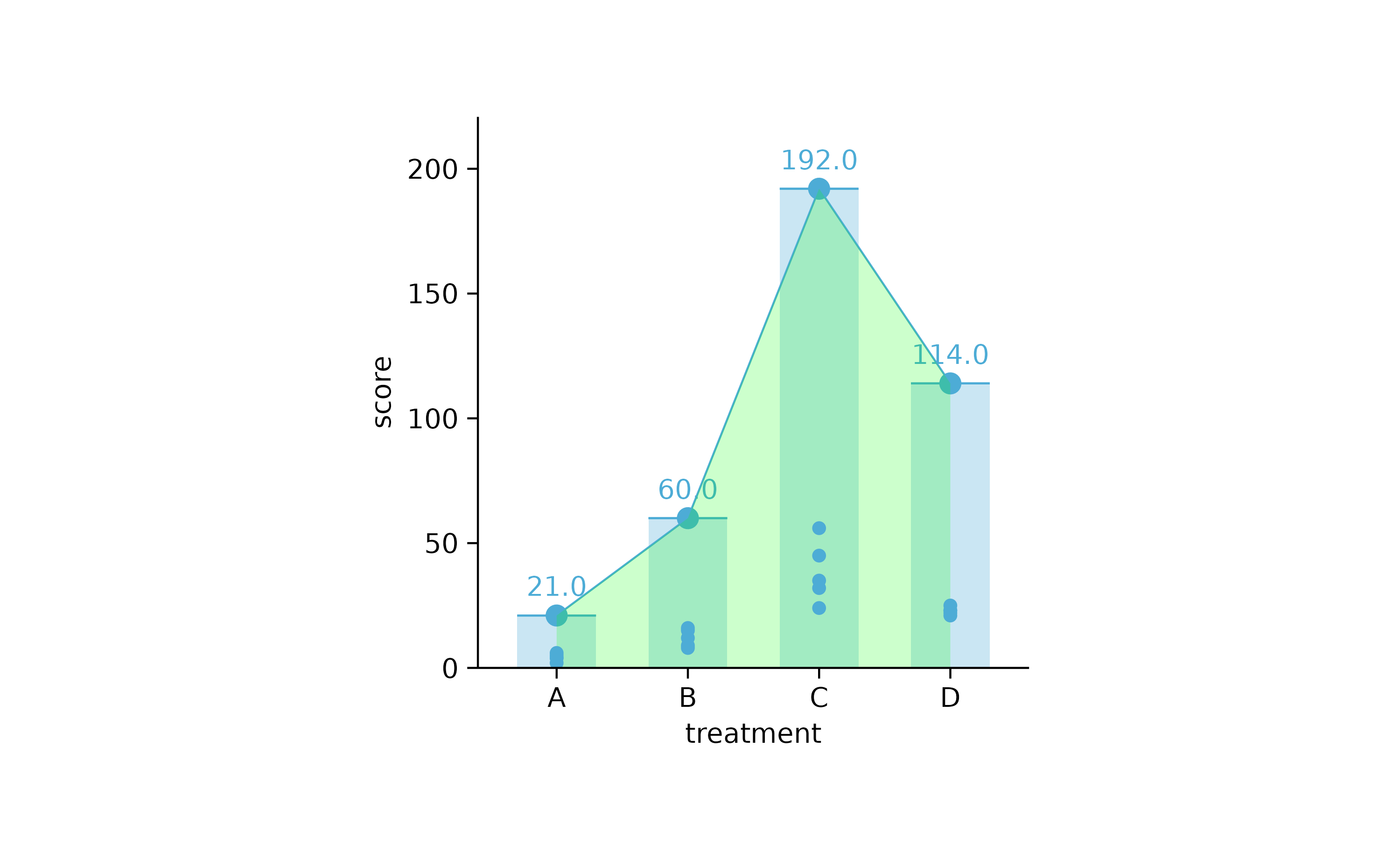
study %>%
tidyplot(score, treatment) %>%
add_sum_bar(alpha = 0.3) %>%
add_sum_dash() %>%
add_sum_dot() %>%
add_sum_value(extra_padding = 0.3) %>%
add_sum_line() %>%
add_sum_area(alpha = 0.2, fill = "green") %>%
add_data_points()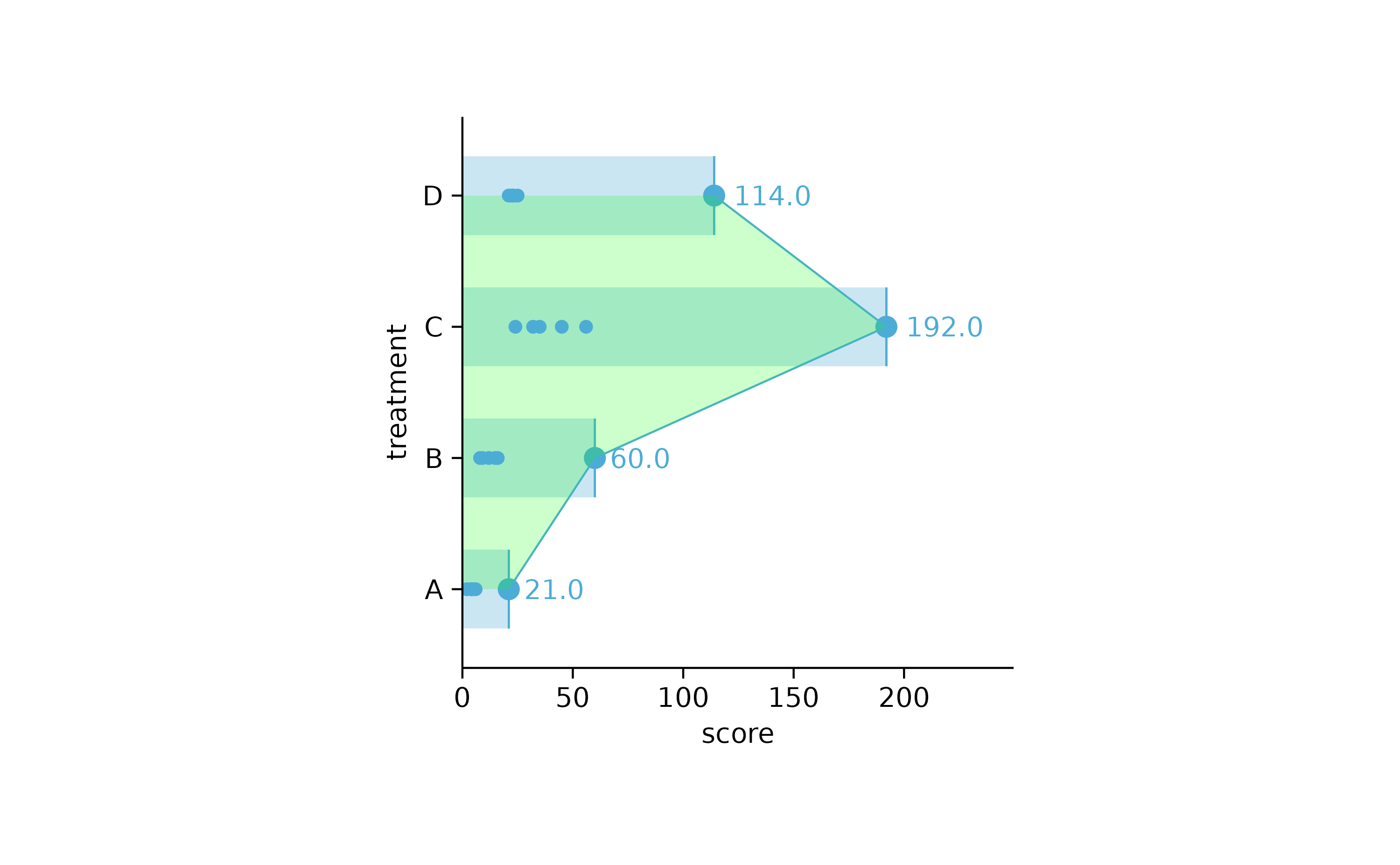
study %>%
tidyplot(treatment) %>%
add_count_bar(alpha = 0.3) %>%
add_count_dash() %>%
add_count_dot() %>%
add_count_value() %>%
add_count_line() %>%
add_count_area(alpha = 0.2, fill = "green")
# grouped
study %>%
tidyplot(group, score, color = dose) %>%
add_error_bar() %>%
add_sd_bar() %>%
add_range_bar() %>%
add_ci95_bar() %>%
add_data_points()
study %>%
tidyplot(group, score, color = dose) %>%
add_error_ribbon() %>%
add_sd_ribbon() %>%
add_range_ribbon() %>%
add_ci95_ribbon() %>%
add_data_points()
study %>%
tidyplot(group, score, color = dose) %>%
add_mean_bar(alpha = 0.3) %>%
add_mean_dash() %>%
add_mean_dot() %>%
add_mean_value() %>%
add_mean_line() %>%
add_mean_area(alpha = 0.5) %>%
add_data_points()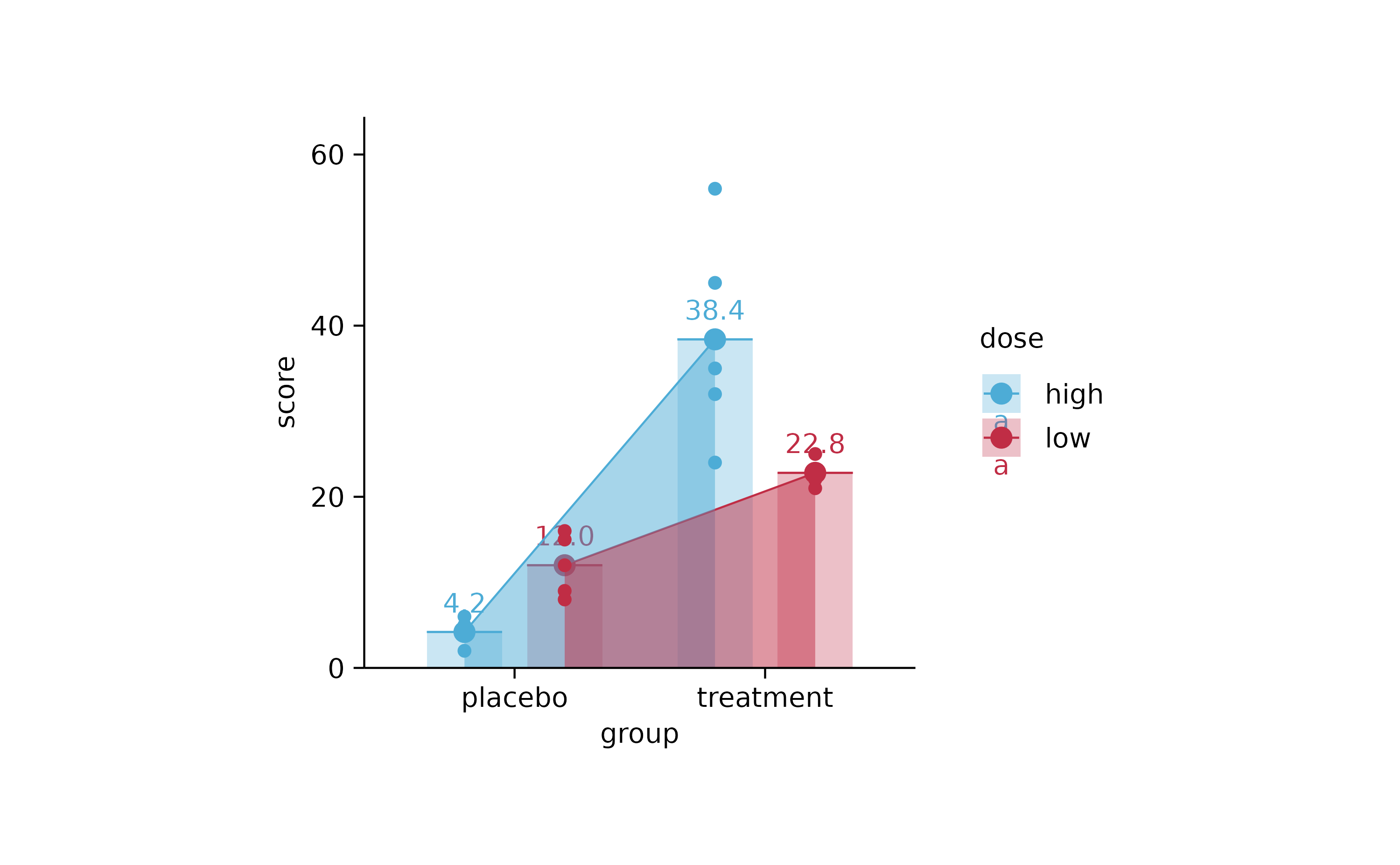
study %>%
tidyplot(group, score, color = dose) %>%
add_median_bar(alpha = 0.3) %>%
add_median_dash() %>%
add_median_dot() %>%
add_median_value() %>%
add_median_line() %>%
add_median_area(alpha = 0.5) %>%
add_data_points()
study %>%
tidyplot(group, score, color = dose) %>%
add_sum_bar(alpha = 0.3) %>%
add_sum_dash() %>%
add_sum_dot() %>%
add_sum_value() %>%
add_sum_line() %>%
add_sum_area(alpha = 0.5) %>%
add_data_points()
study %>%
tidyplot(group, color = dose) %>%
add_count_bar(alpha = 0.3) %>%
add_count_dash() %>%
add_count_dot() %>%
add_count_value() %>%
add_count_line() %>%
add_count_area(alpha = 0.5)
# add_data_points()
animals %>%
tidyplot(x = weight, y = size, color = number_of_legs) %>%
add_data_points()
animals %>%
tidyplot(x = weight, y = size, color = number_of_legs) %>%
add_data_points(confetti = TRUE)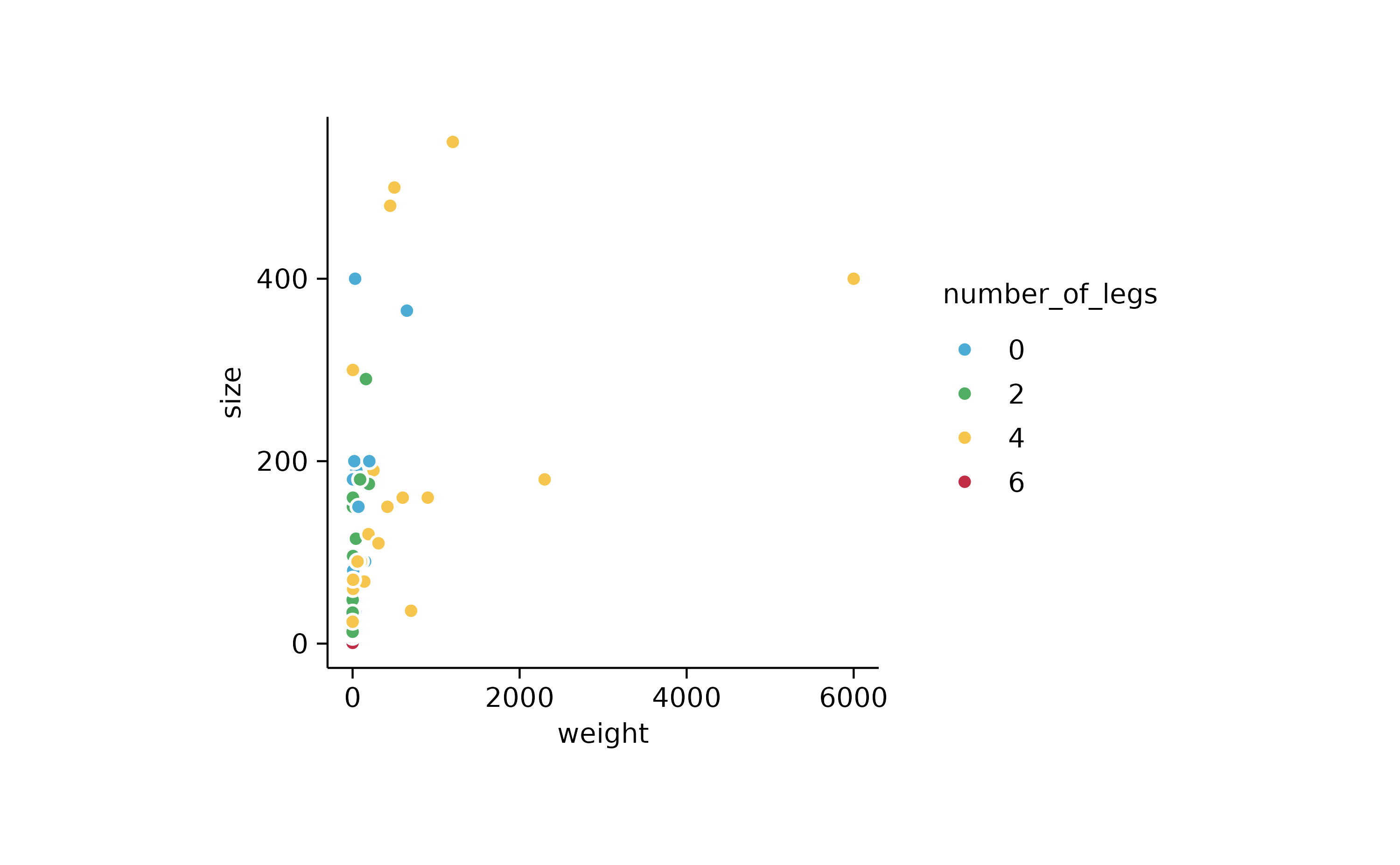
## x is discrete
study %>%
tidyplot(x = treatment, y = score, color = treatment) %>%
add_data_points()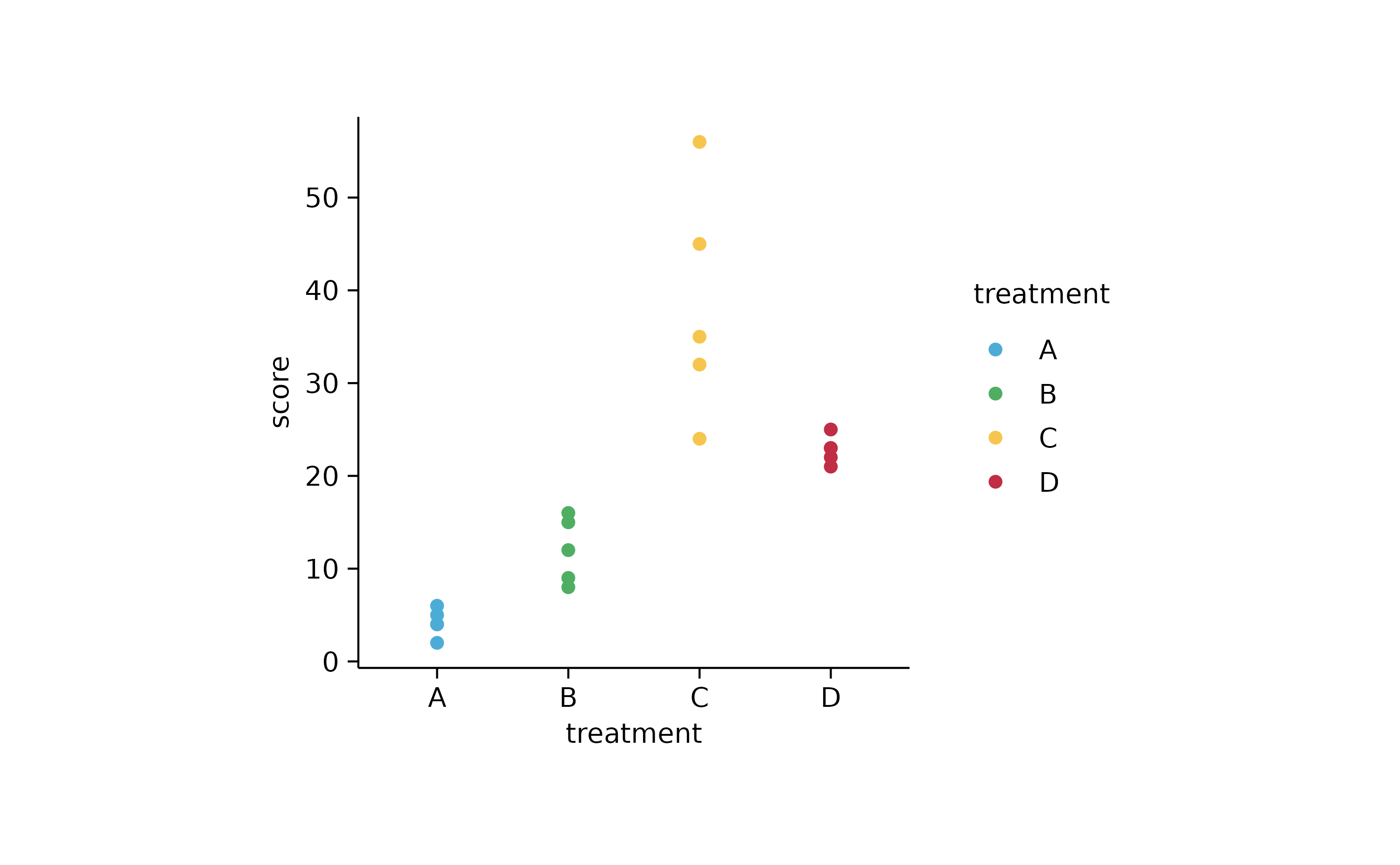
study %>%
tidyplot(x = treatment, y = score, color = treatment) %>%
add_data_points_jitter()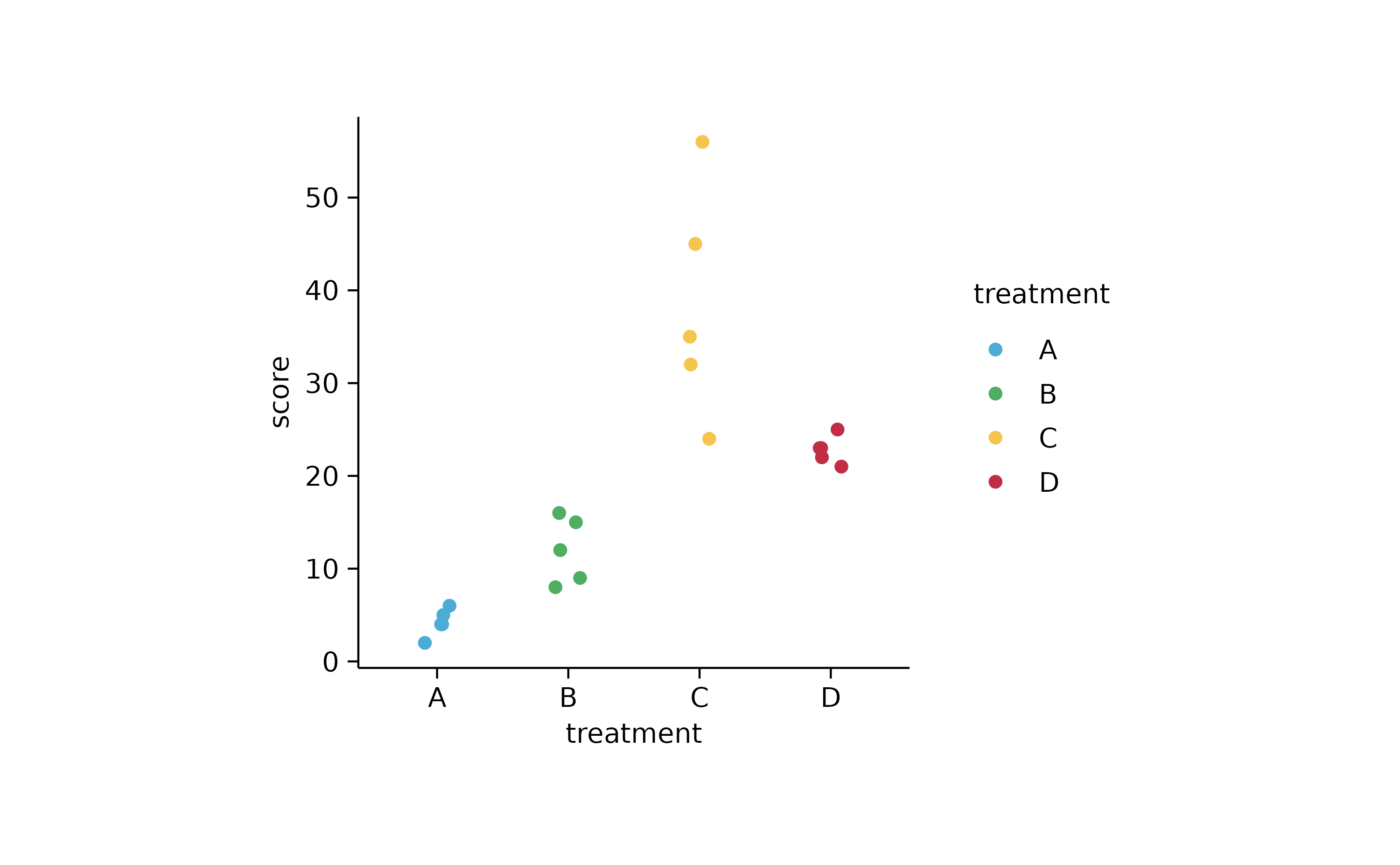
study %>%
tidyplot(x = treatment, y = score, color = treatment) %>%
add_data_points_beeswarm()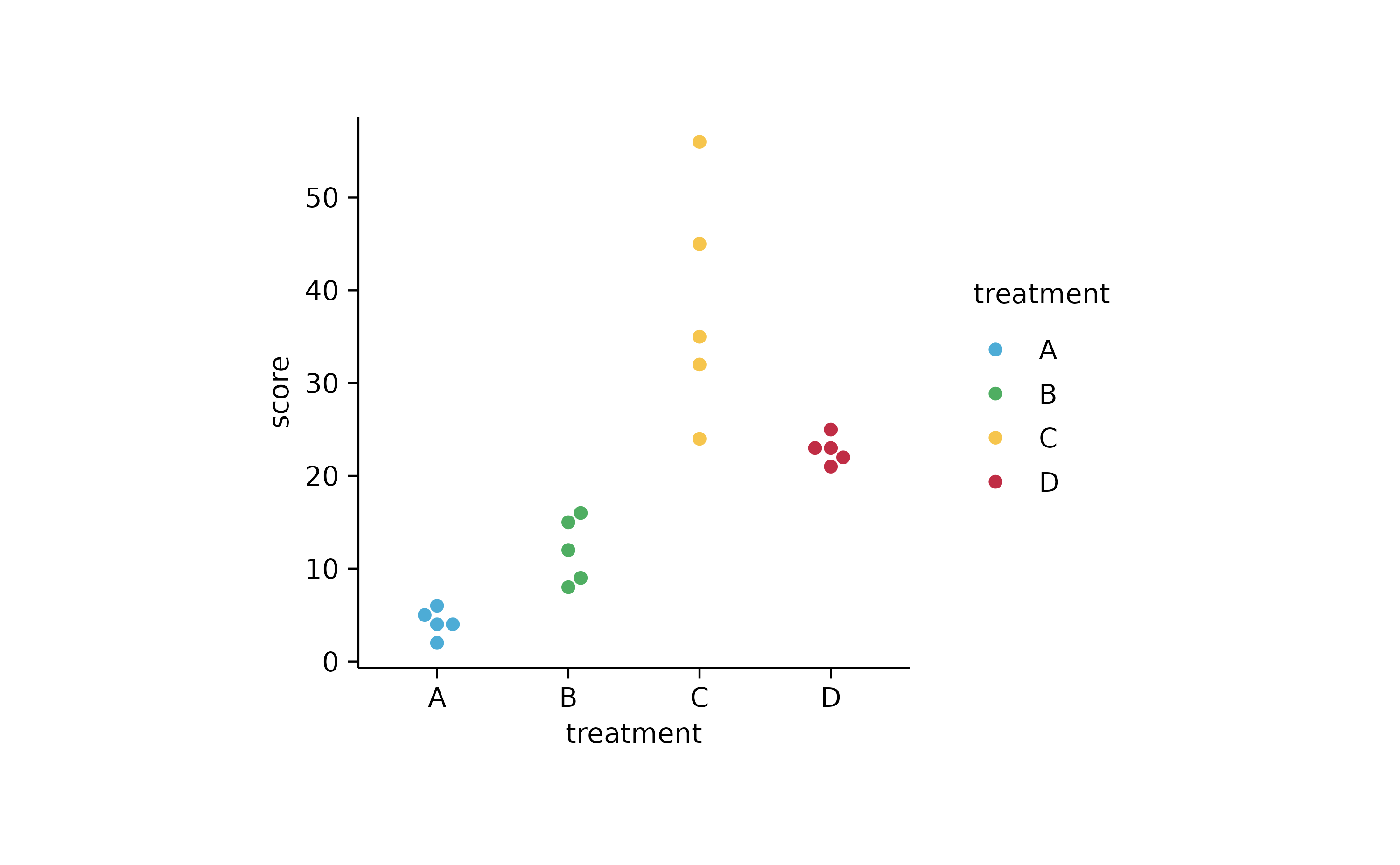
# confetti
study %>%
tidyplot(x = treatment, y = score, color = treatment) %>%
add_data_points(confetti = TRUE)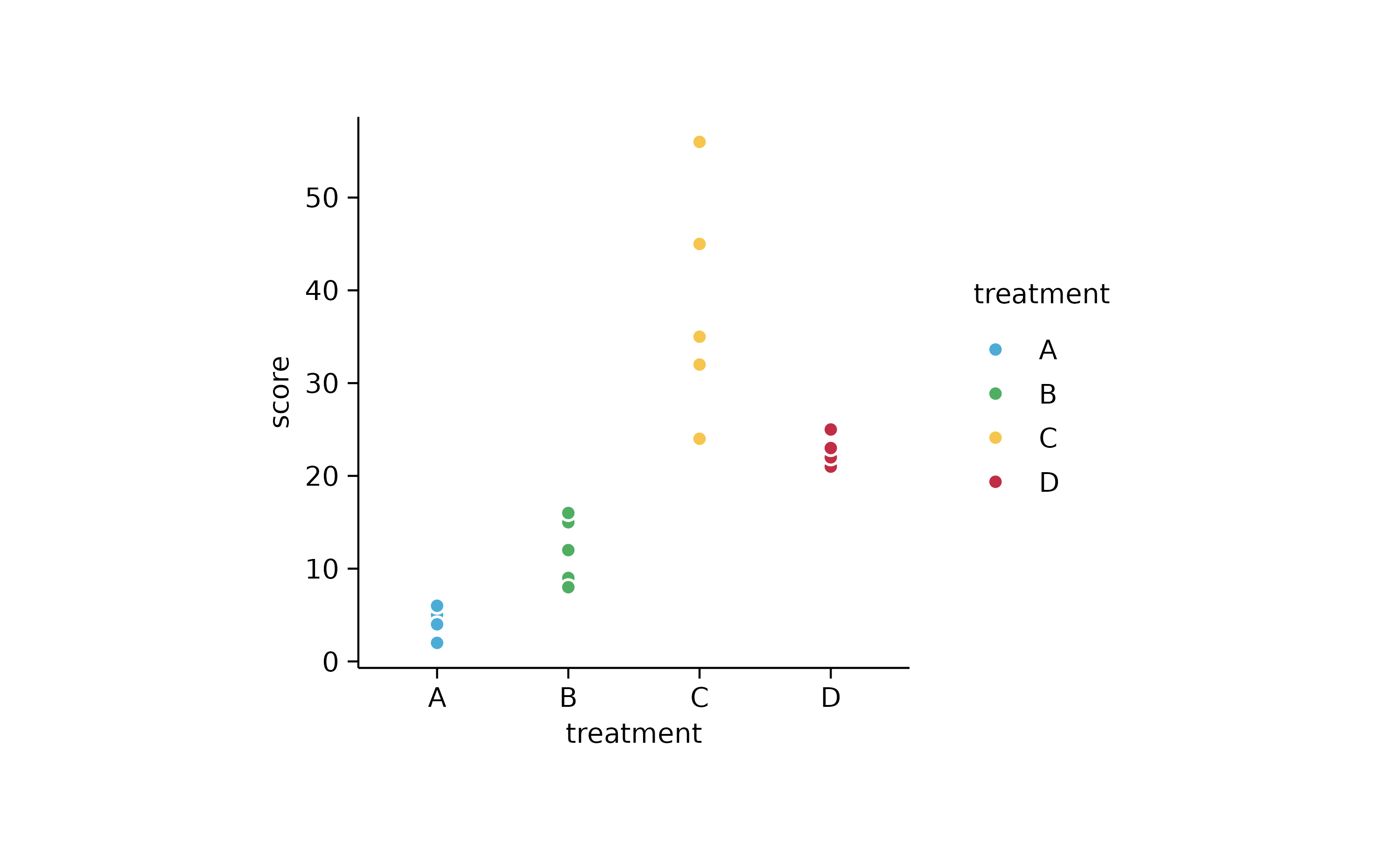
study %>%
tidyplot(x = treatment, y = score, color = treatment) %>%
add_data_points_jitter(confetti = TRUE)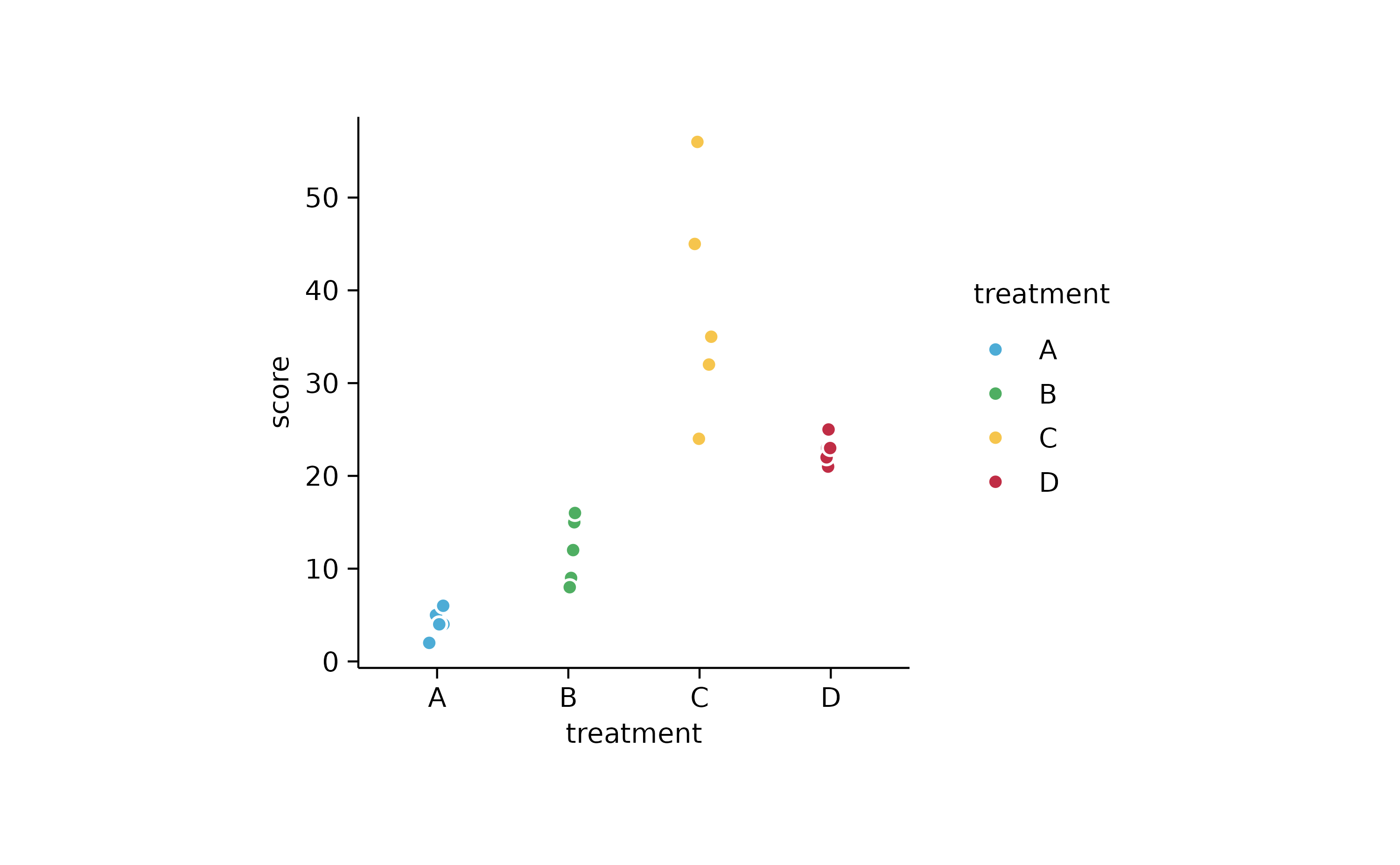
study %>%
tidyplot(x = treatment, y = score, color = treatment) %>%
add_data_points_beeswarm(confetti = TRUE)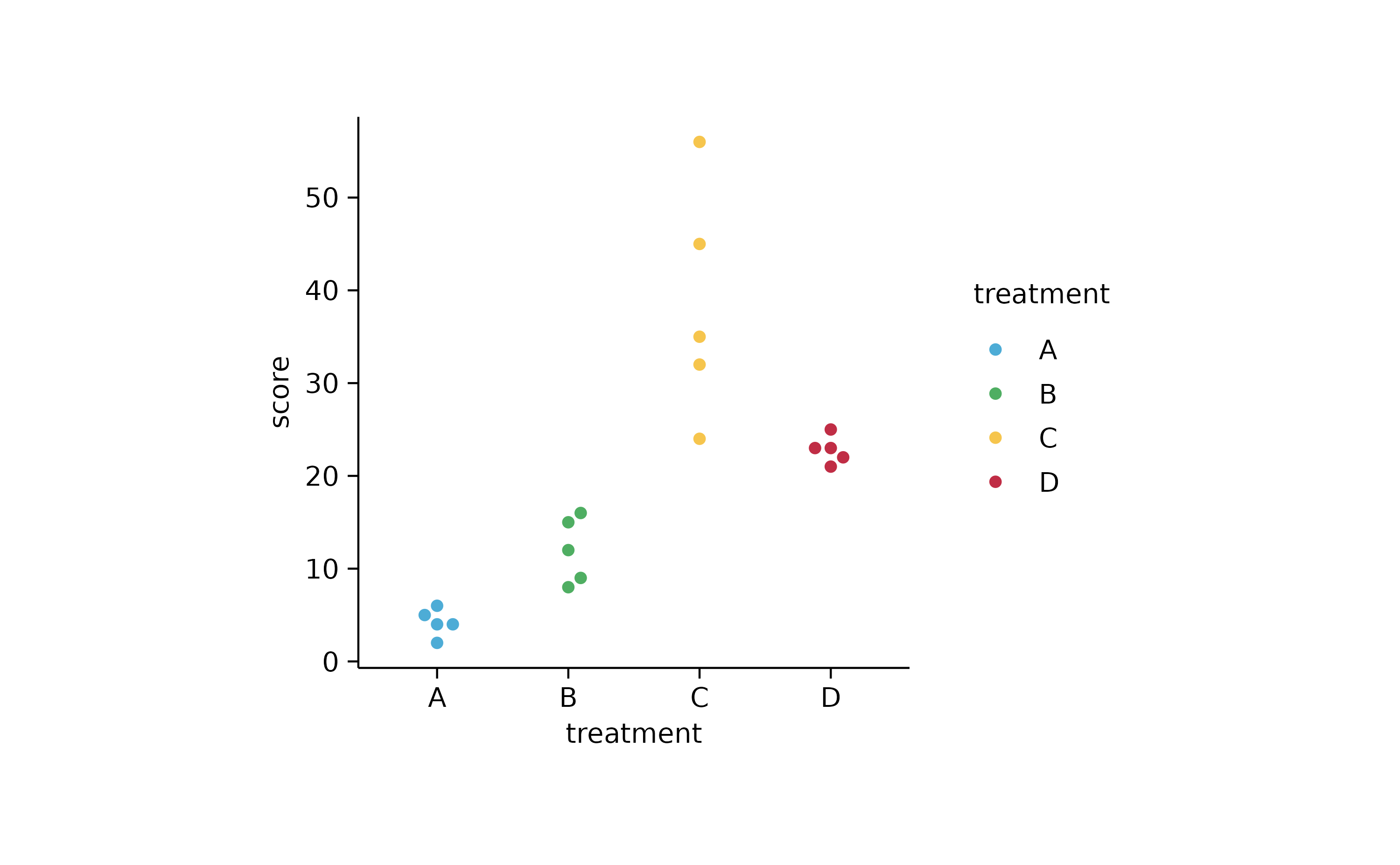
## highlighting
animals %>%
tidyplot(x = weight, y = size) %>%
add_data_points() %>%
add_data_points(data = max_rows(weight, n = 3), color = "red")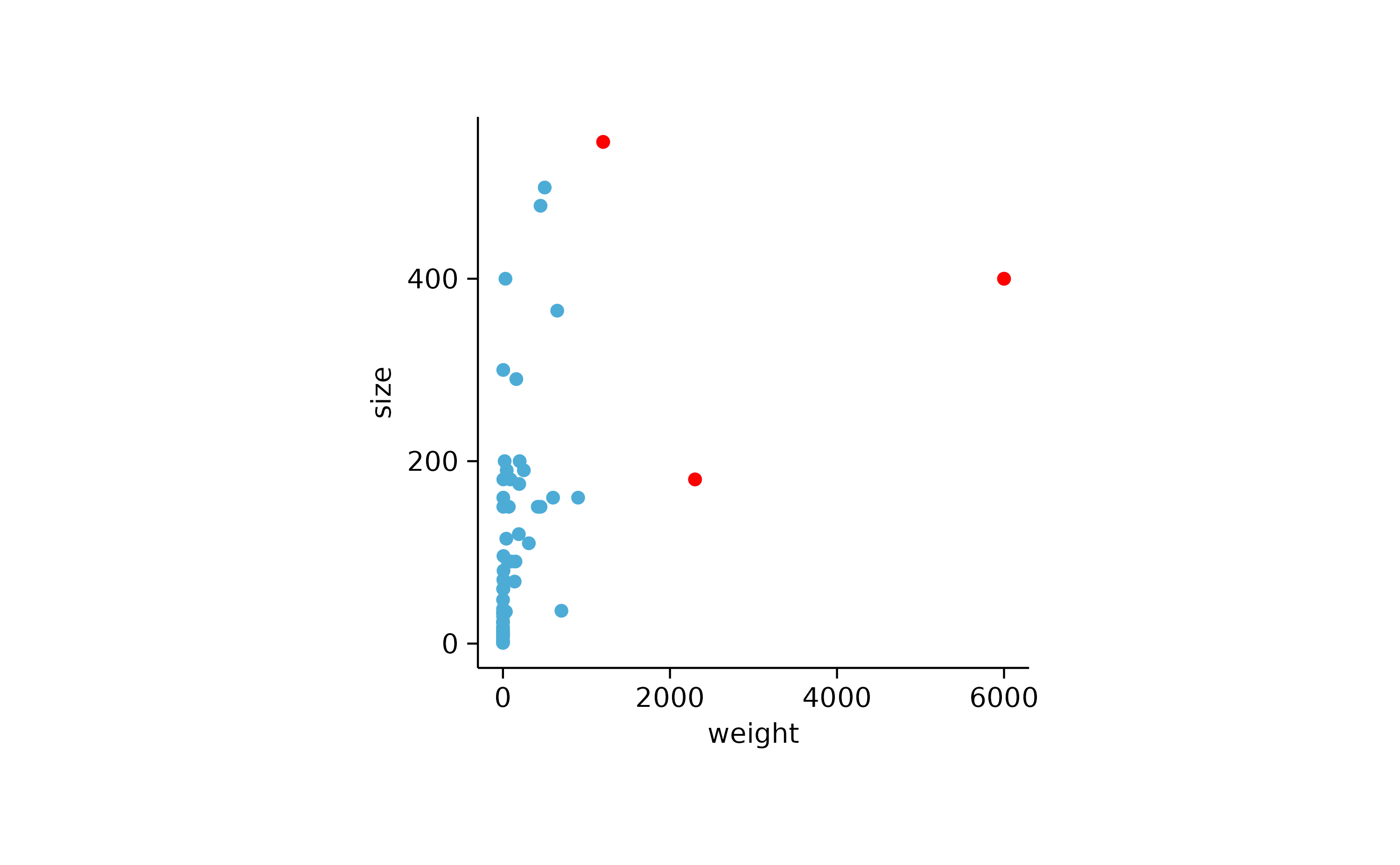
animals %>%
tidyplot(x = weight, y = size) %>%
add_data_points() %>%
add_data_points(data = max_rows(weight, n = 3), color = "red", shape = 1, size = 3)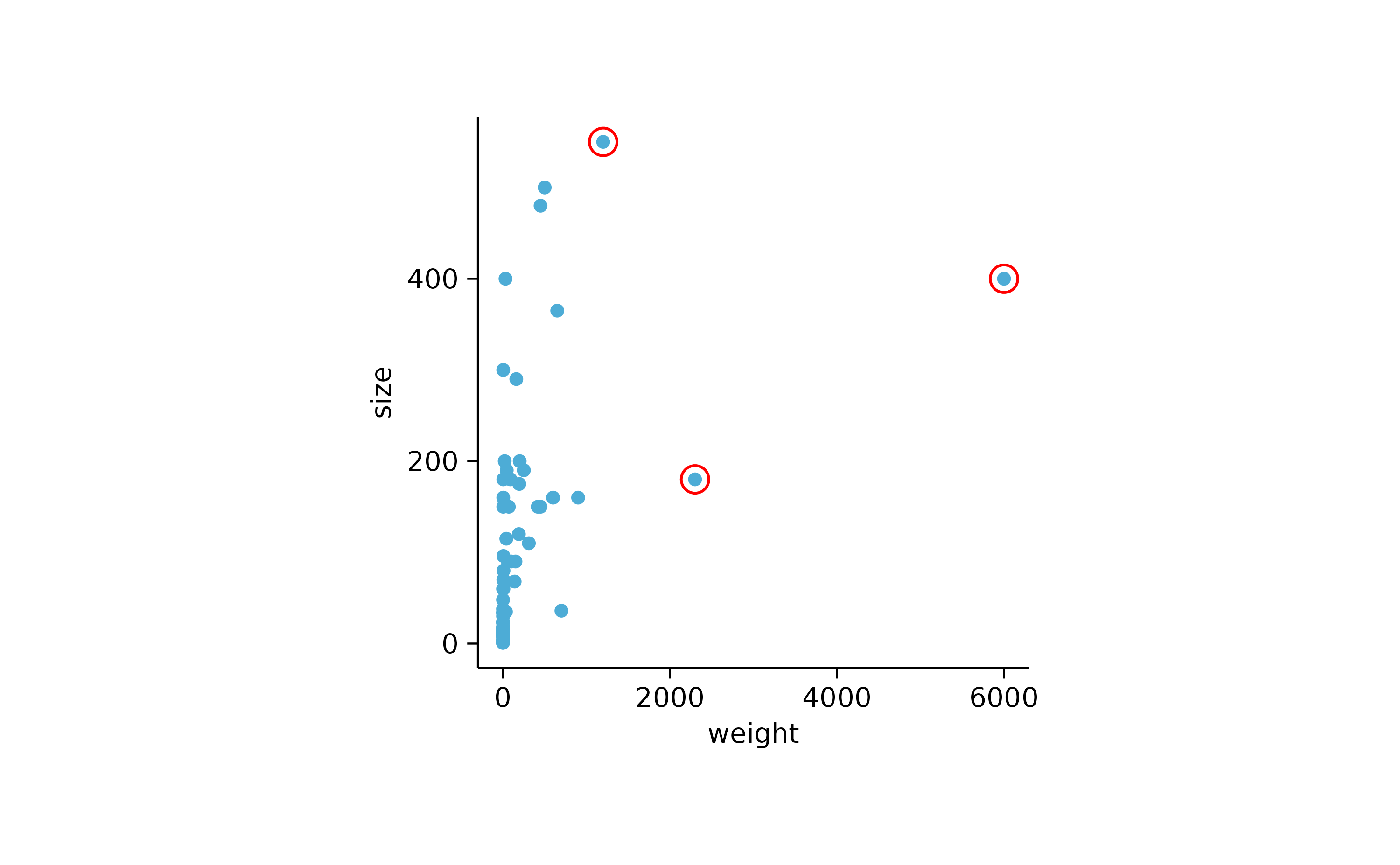
animals %>%
tidyplot(color = family) %>%
add_barstack_absolute(alpha = 0.3)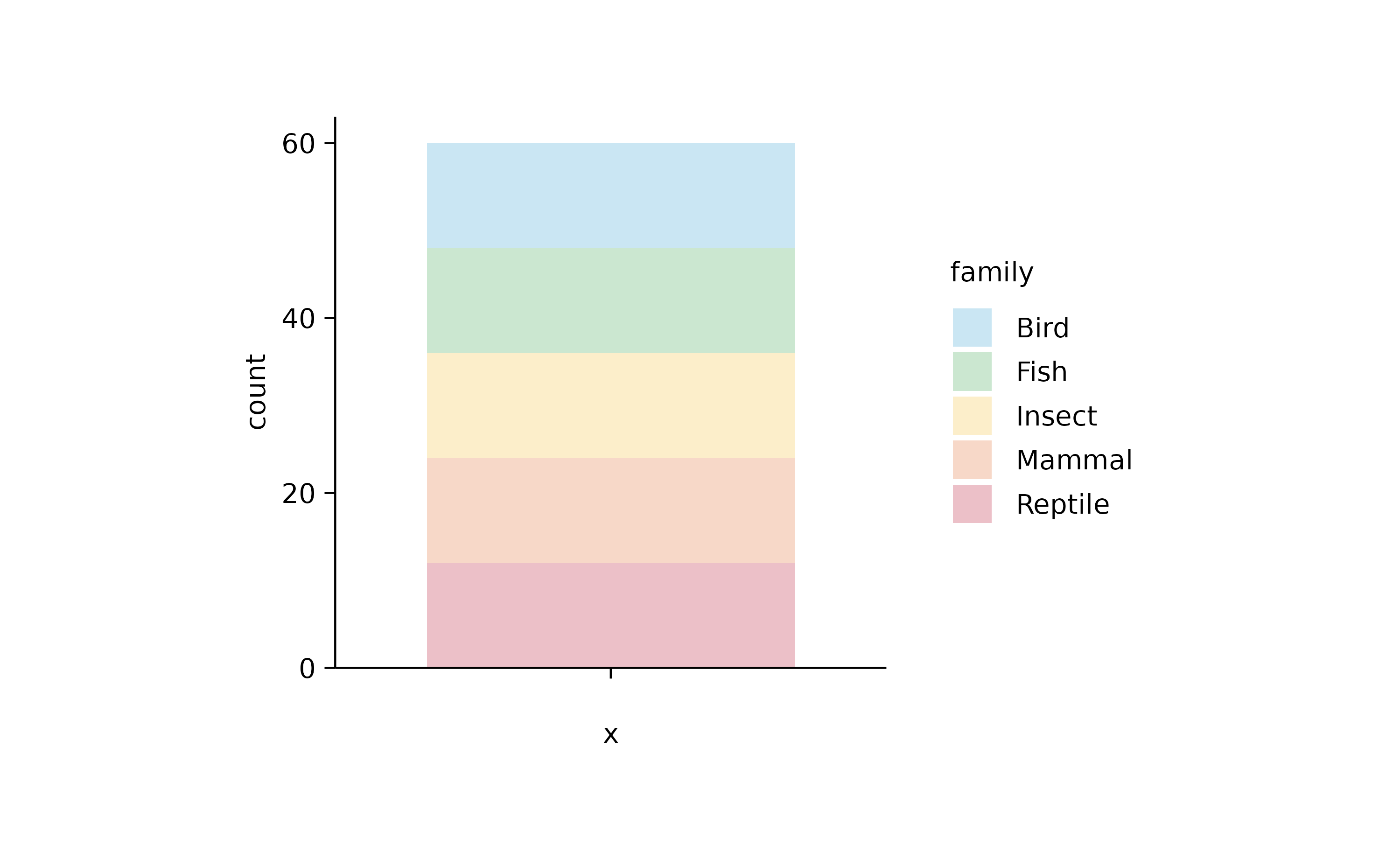
animals %>%
tidyplot(x = diet, color = family) %>%
add_barstack_absolute(alpha = 0.3)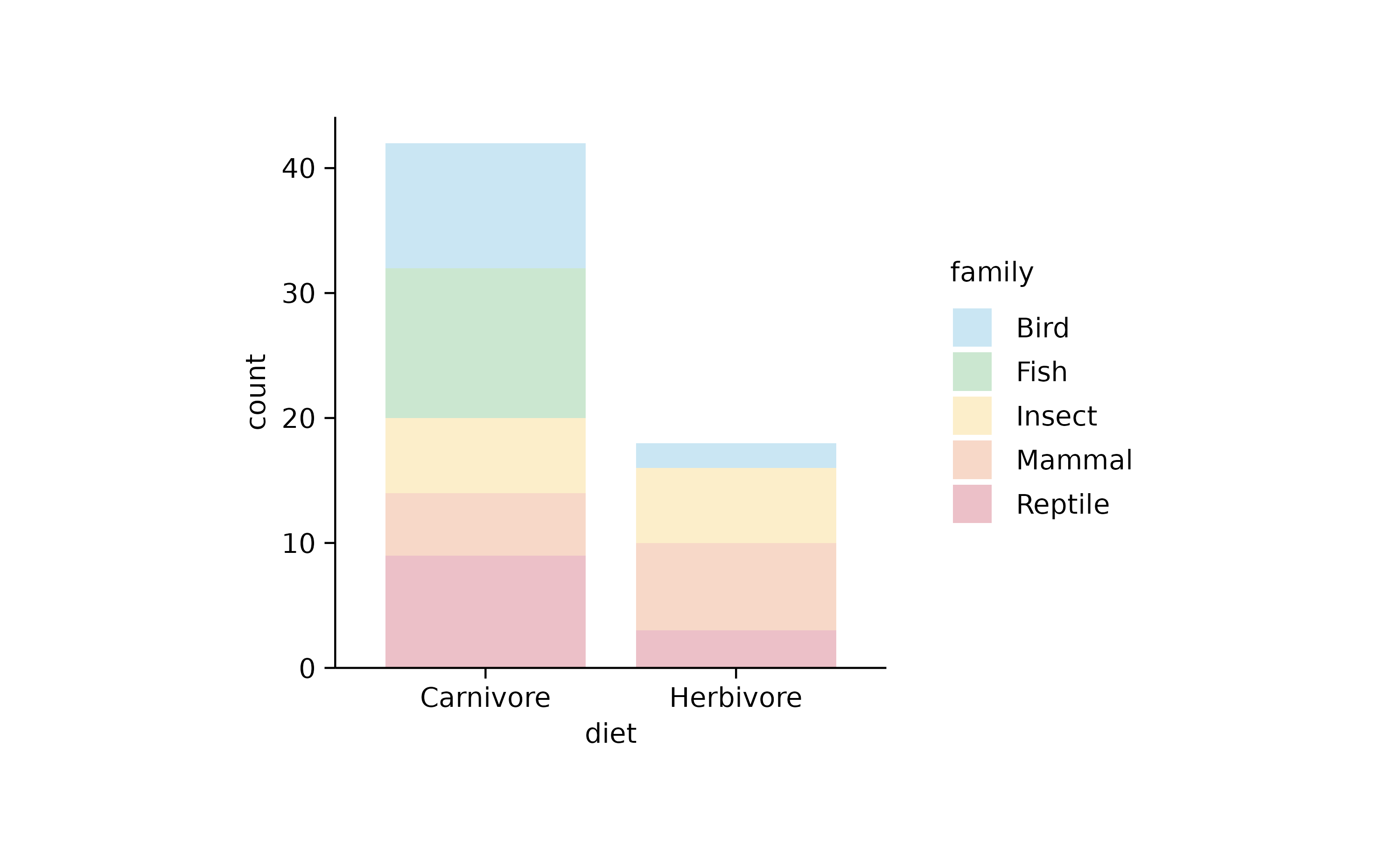
animals %>%
tidyplot(y = diet, color = family) %>%
add_barstack_absolute(alpha = 0.3)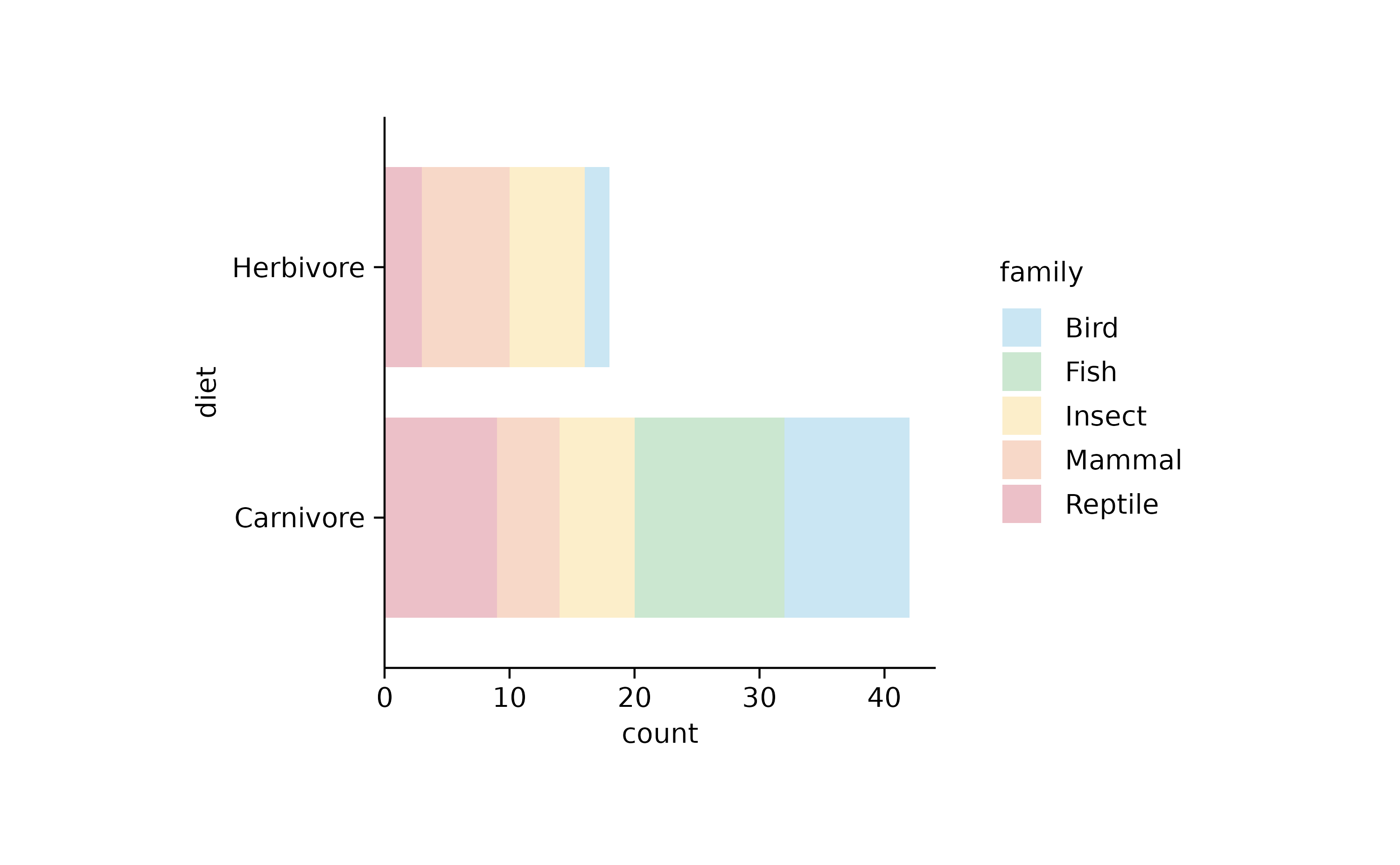
animals %>%
tidyplot(x = diet, y = speed, color = family) %>%
add_barstack_absolute(alpha = 0.3)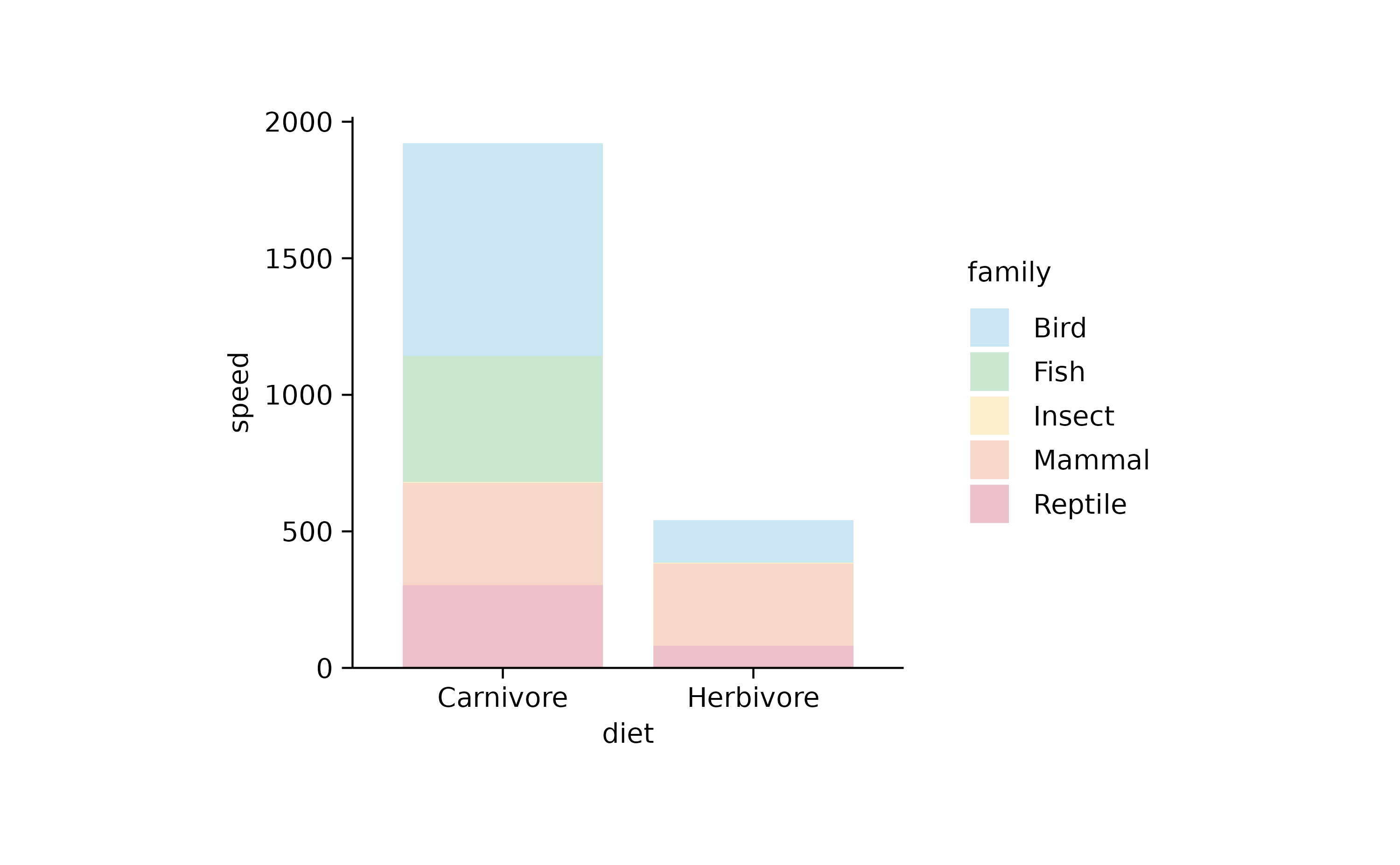
animals %>%
tidyplot(y = diet, x = speed, color = family) %>%
add_barstack_absolute(alpha = 0.3)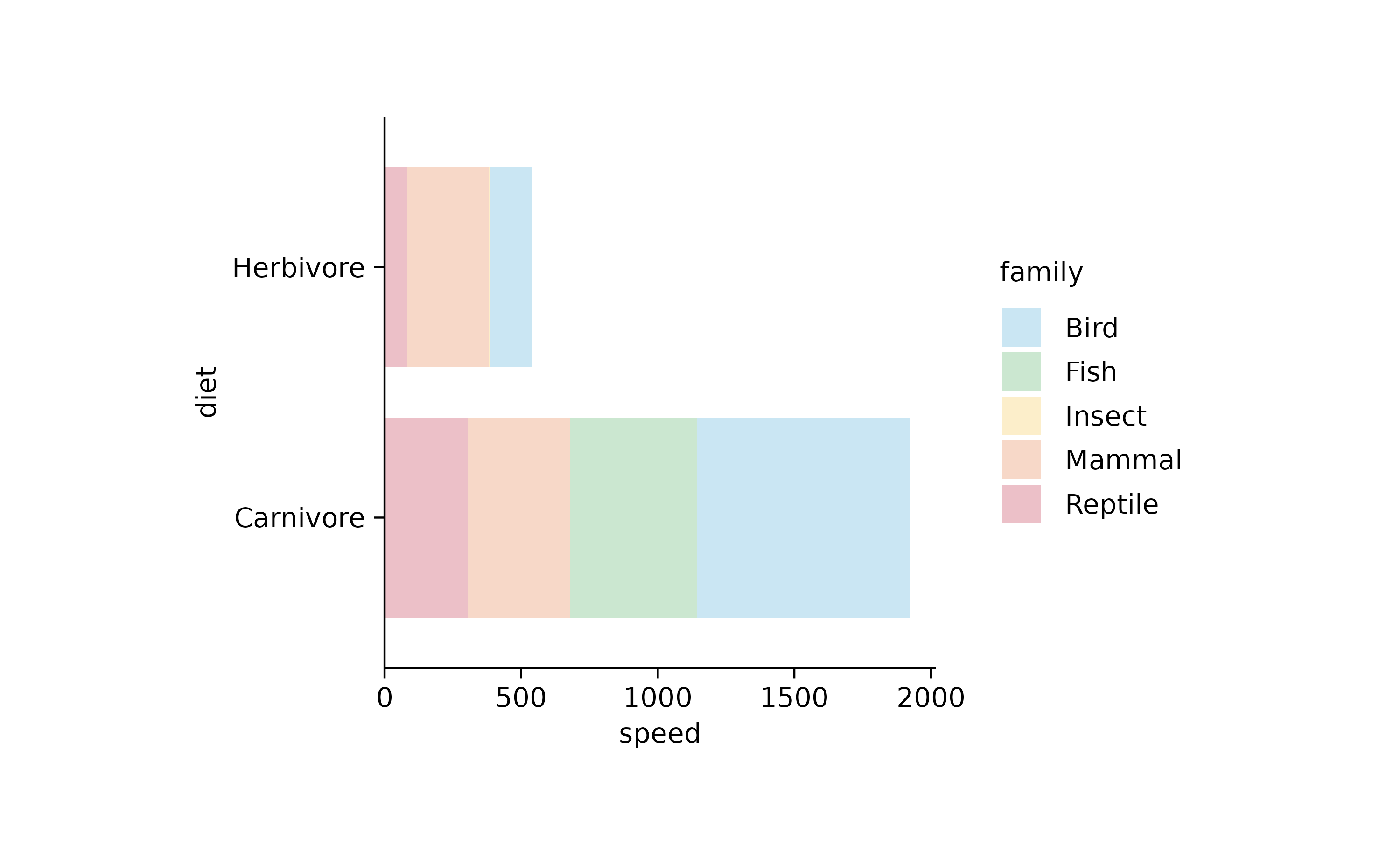
#
animals %>%
tidyplot(color = family) %>%
add_barstack_relative(alpha = 0.3)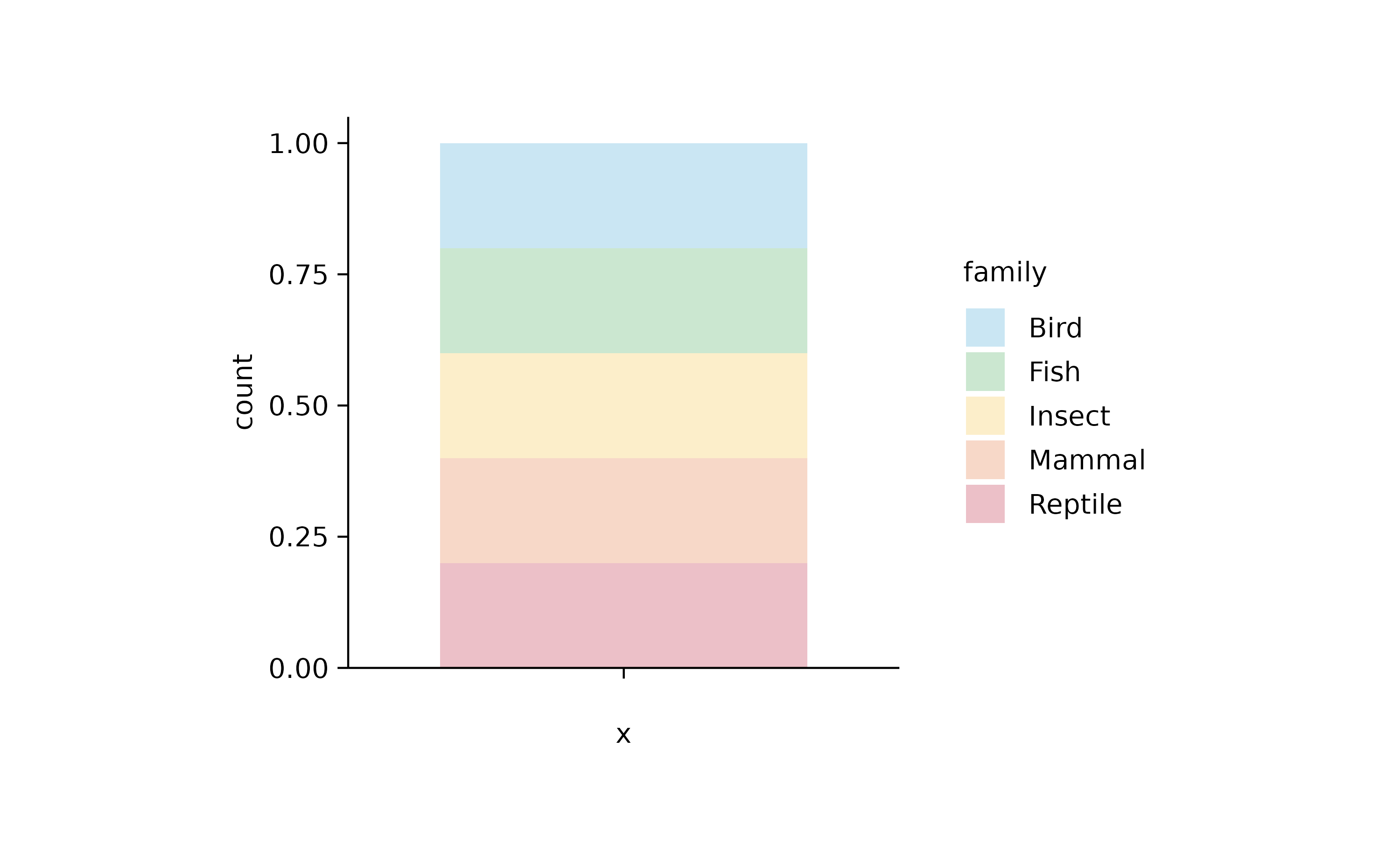
animals %>%
tidyplot(x = diet, color = family) %>%
add_barstack_relative(alpha = 0.3)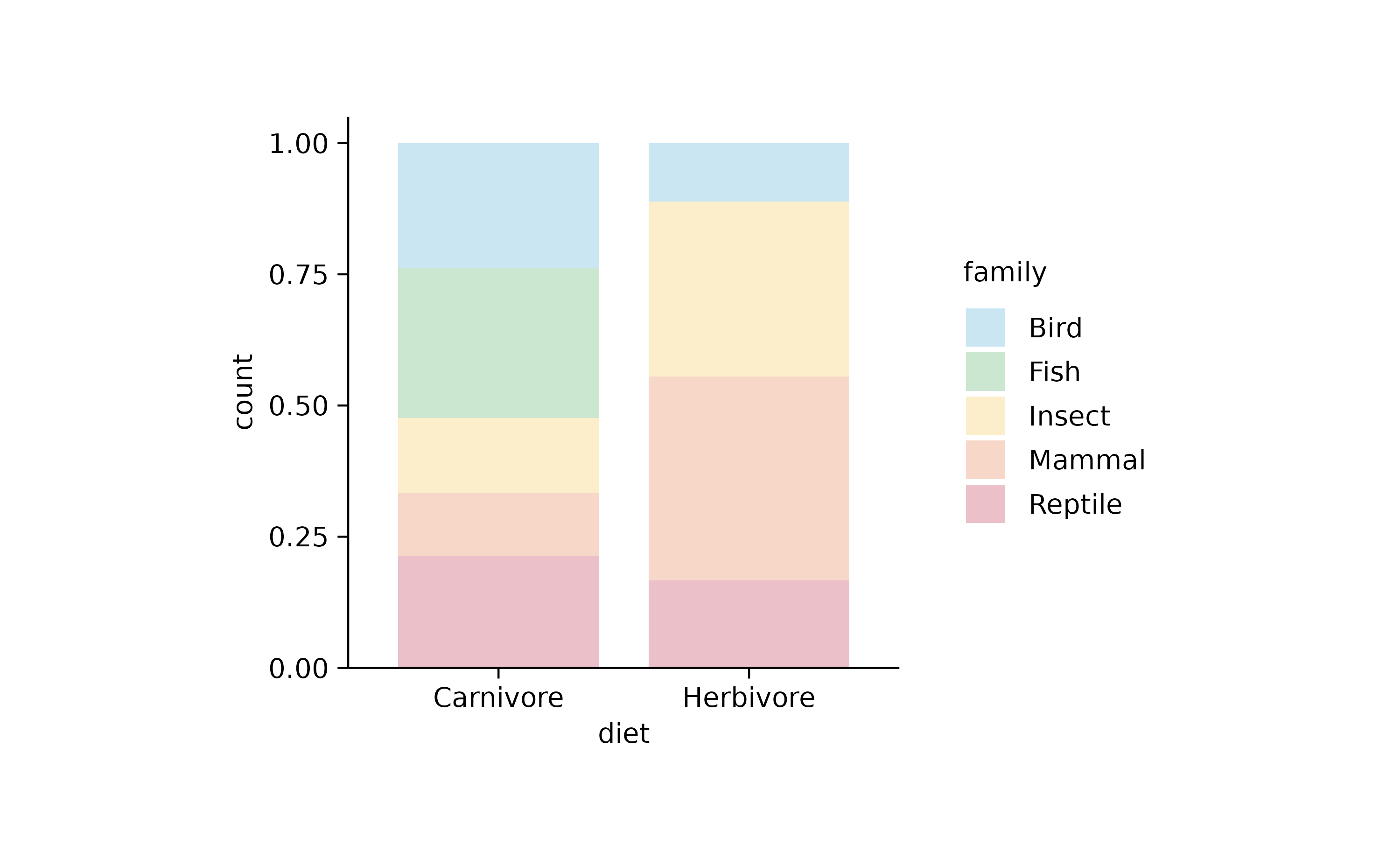
animals %>%
tidyplot(y = diet, color = family) %>%
add_barstack_relative(alpha = 0.3)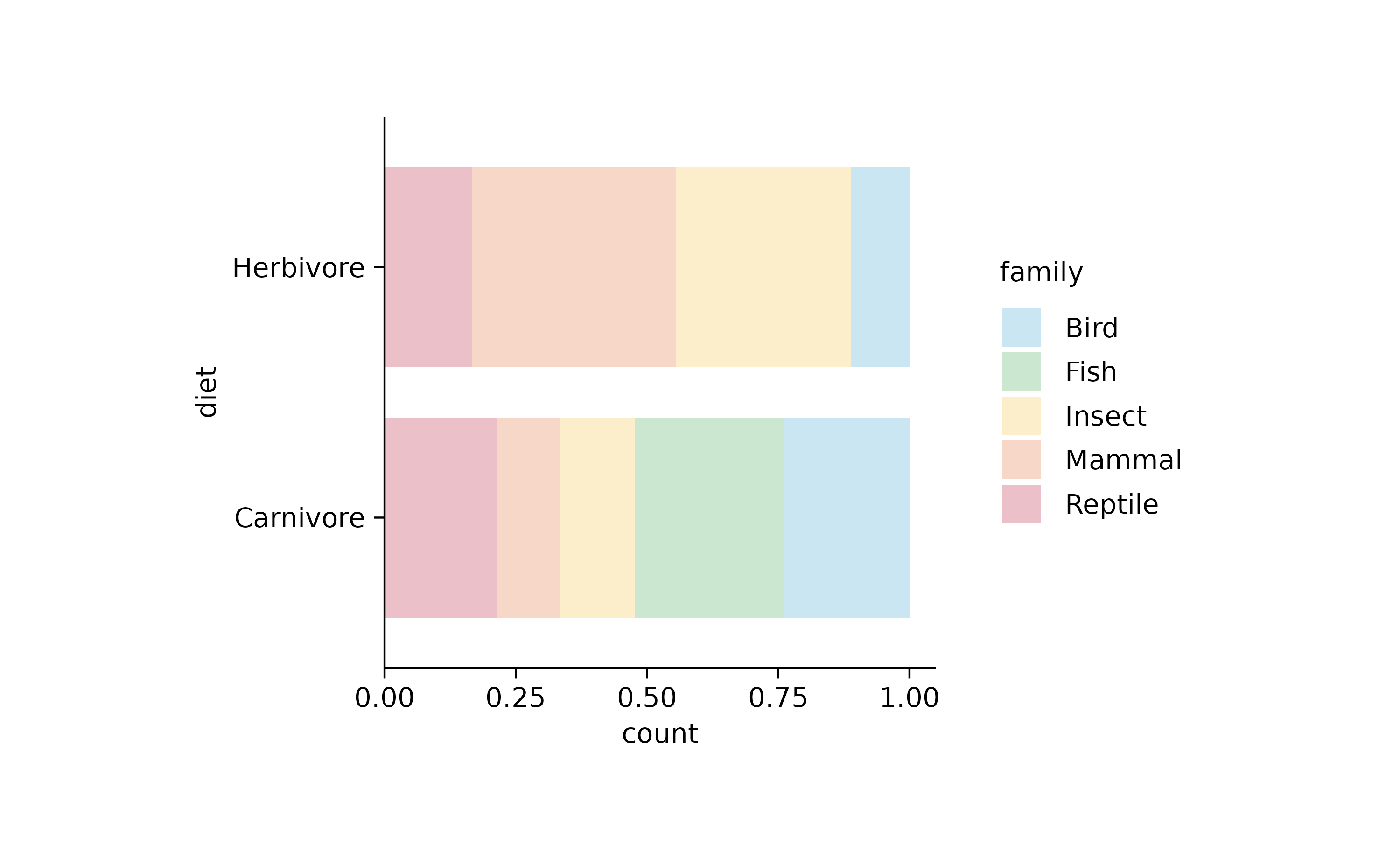
animals %>%
tidyplot(x = diet, y = speed, color = family) %>%
add_barstack_relative(alpha = 0.3)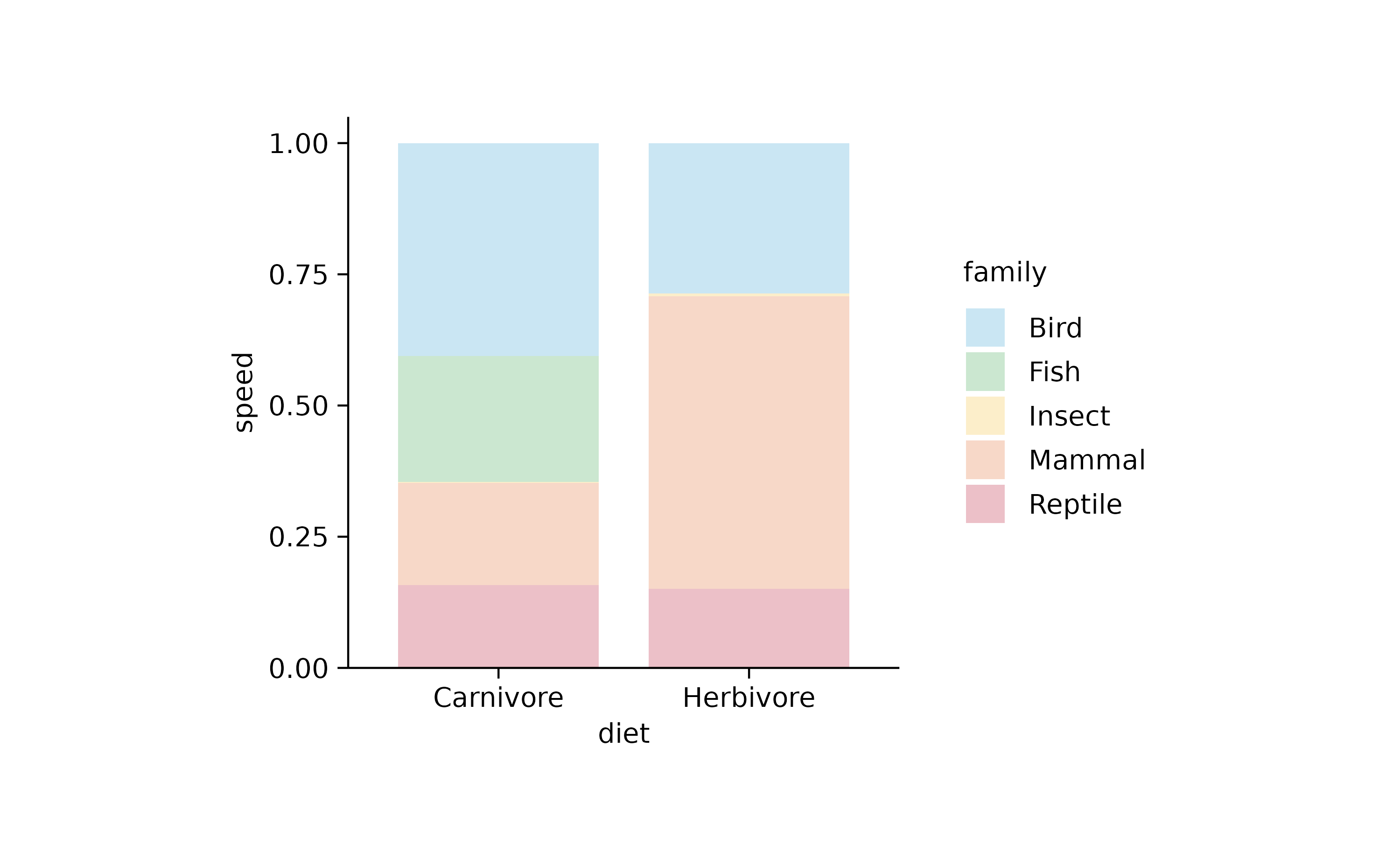
animals %>%
tidyplot(y = diet, x = speed, color = family) %>%
add_barstack_relative(alpha = 0.3)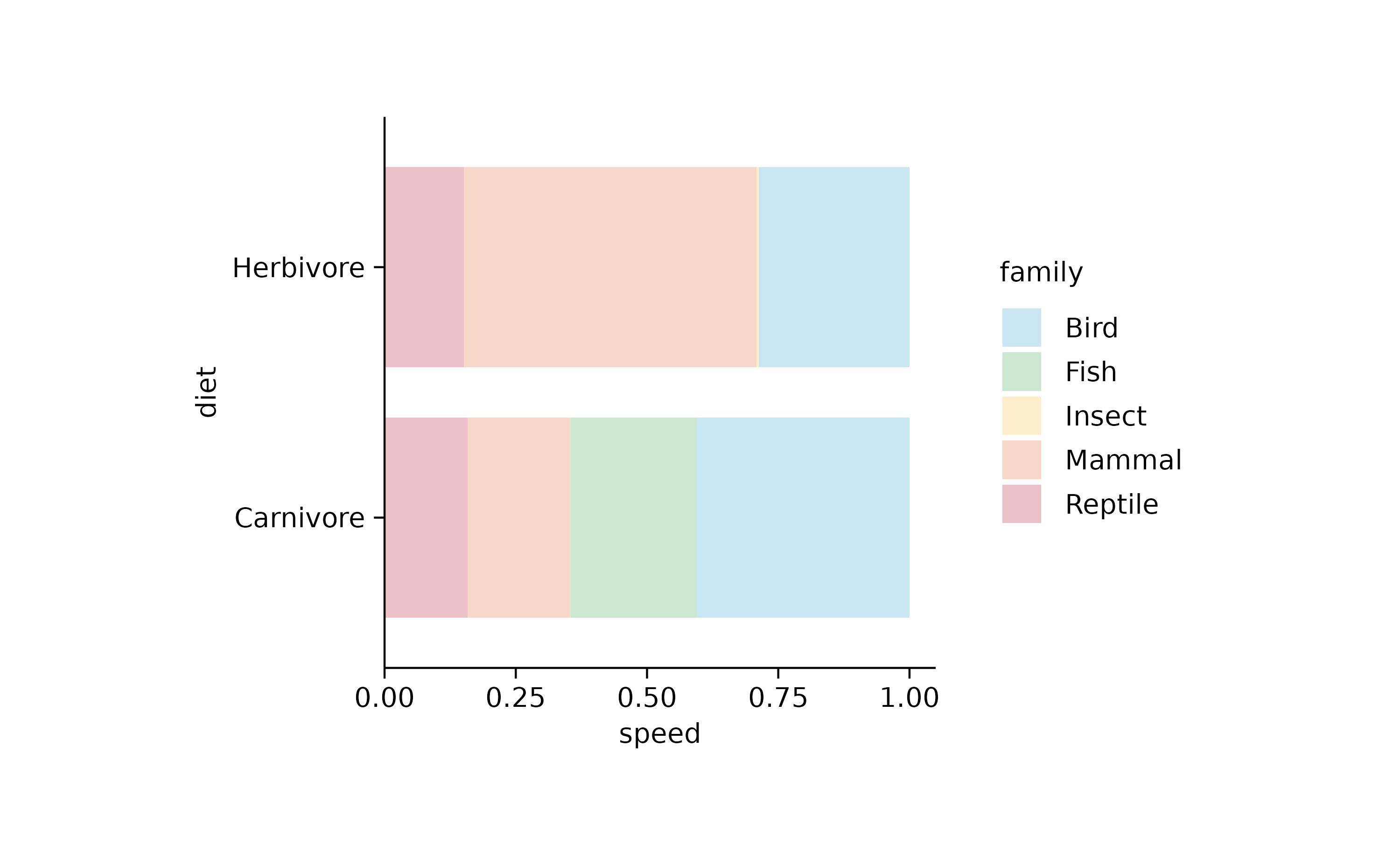
##
animals %>%
tidyplot(x = diet, color = family) %>%
add_areastack_absolute(alpha = 0.3) %>%
reverse_x_axis_labels()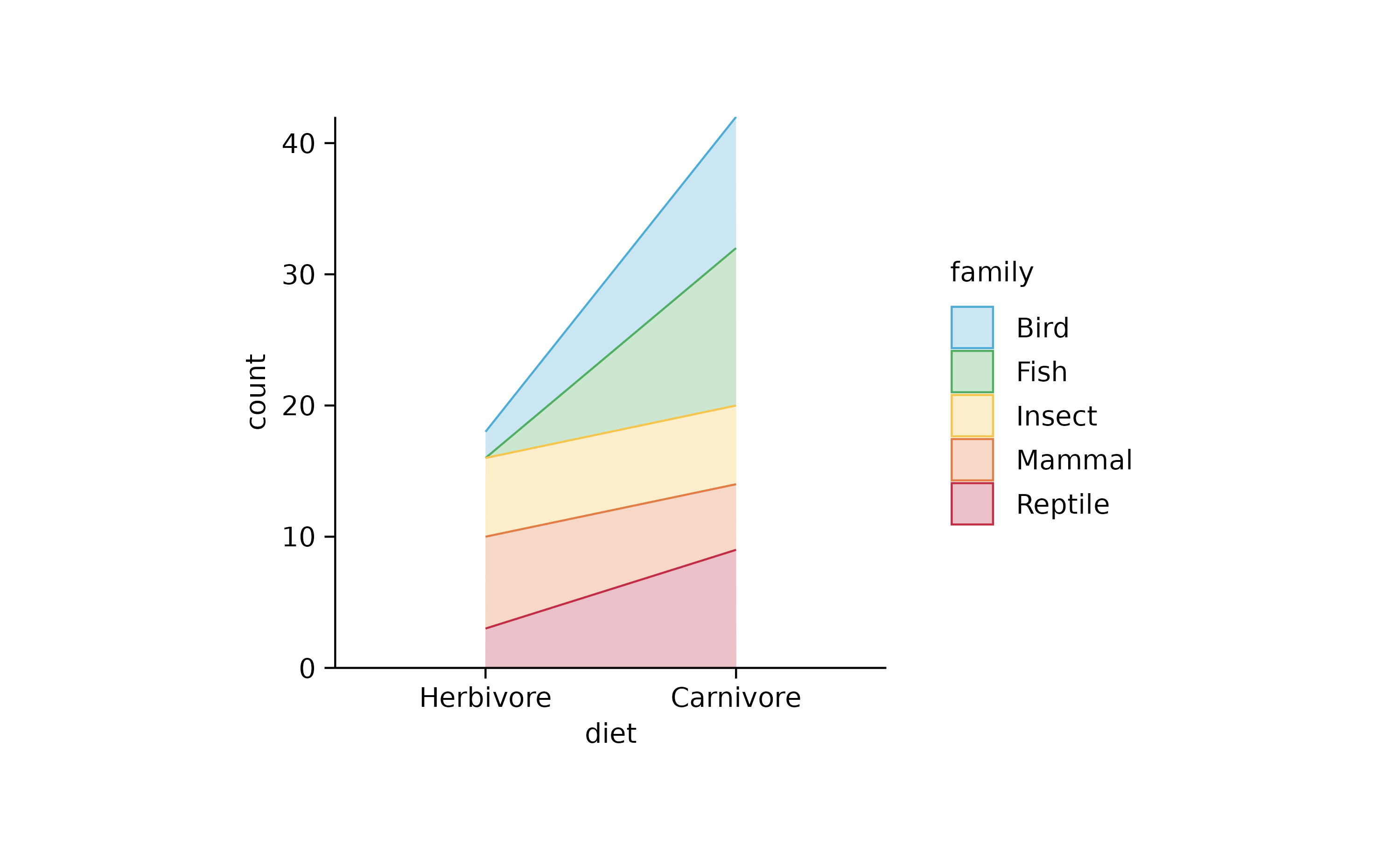
animals %>%
tidyplot(x = diet, y = speed, color = family) %>%
add_areastack_absolute(alpha = 0.3)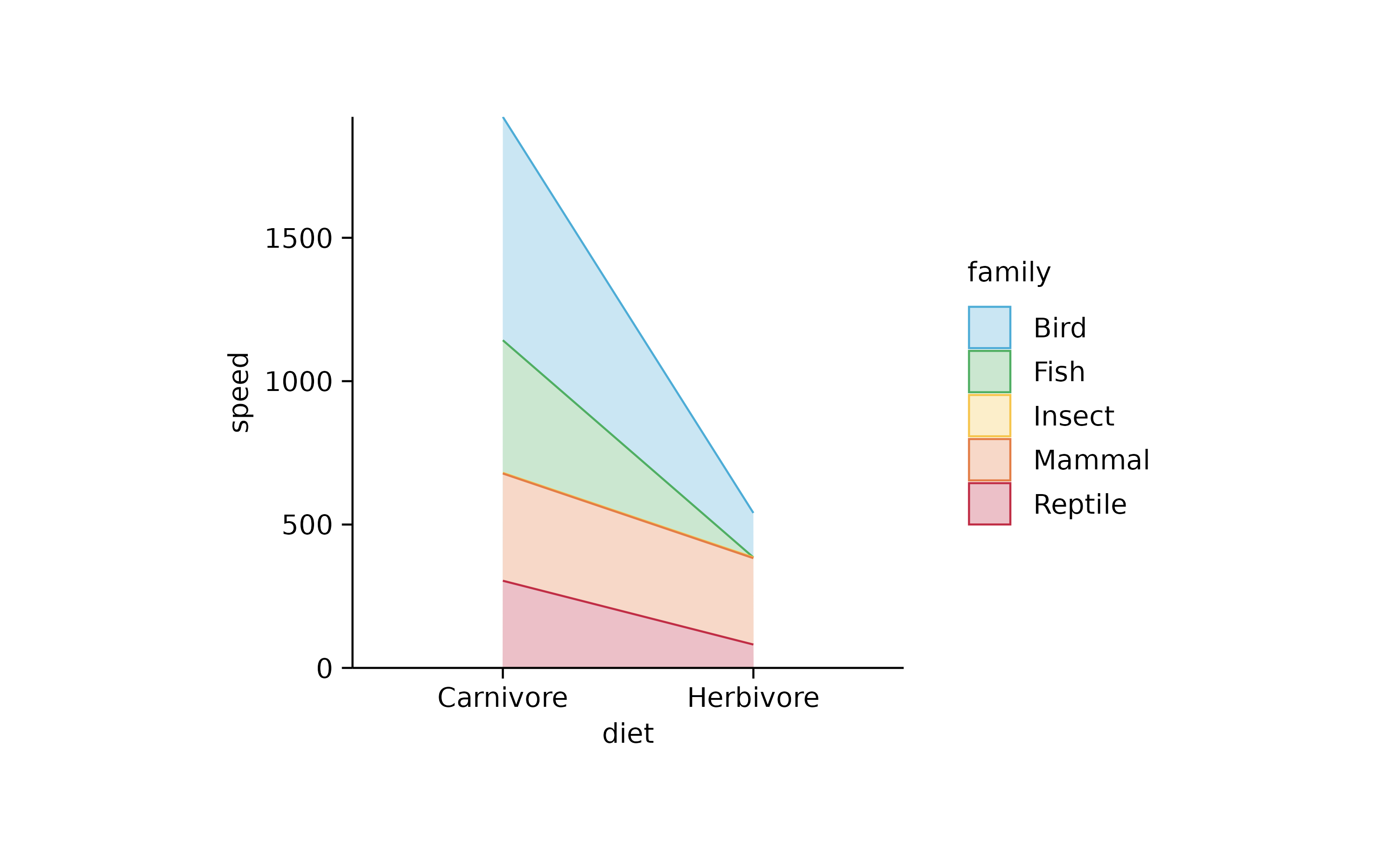
# as_tidyplot
library(ggplot2)
p <-
study %>%
ggplot(aes(x = treatment, y = score, color = treatment, fill = treatment)) +
stat_summary(fun = mean, geom = "bar", color = NA, width = 0.6, alpha = 0.3) +
stat_summary(fun.data = mean_se, geom = "errorbar", linewidth = 0.25, width = 0.4) +
geom_point(position = position_jitterdodge(jitter.width = 0.2))
p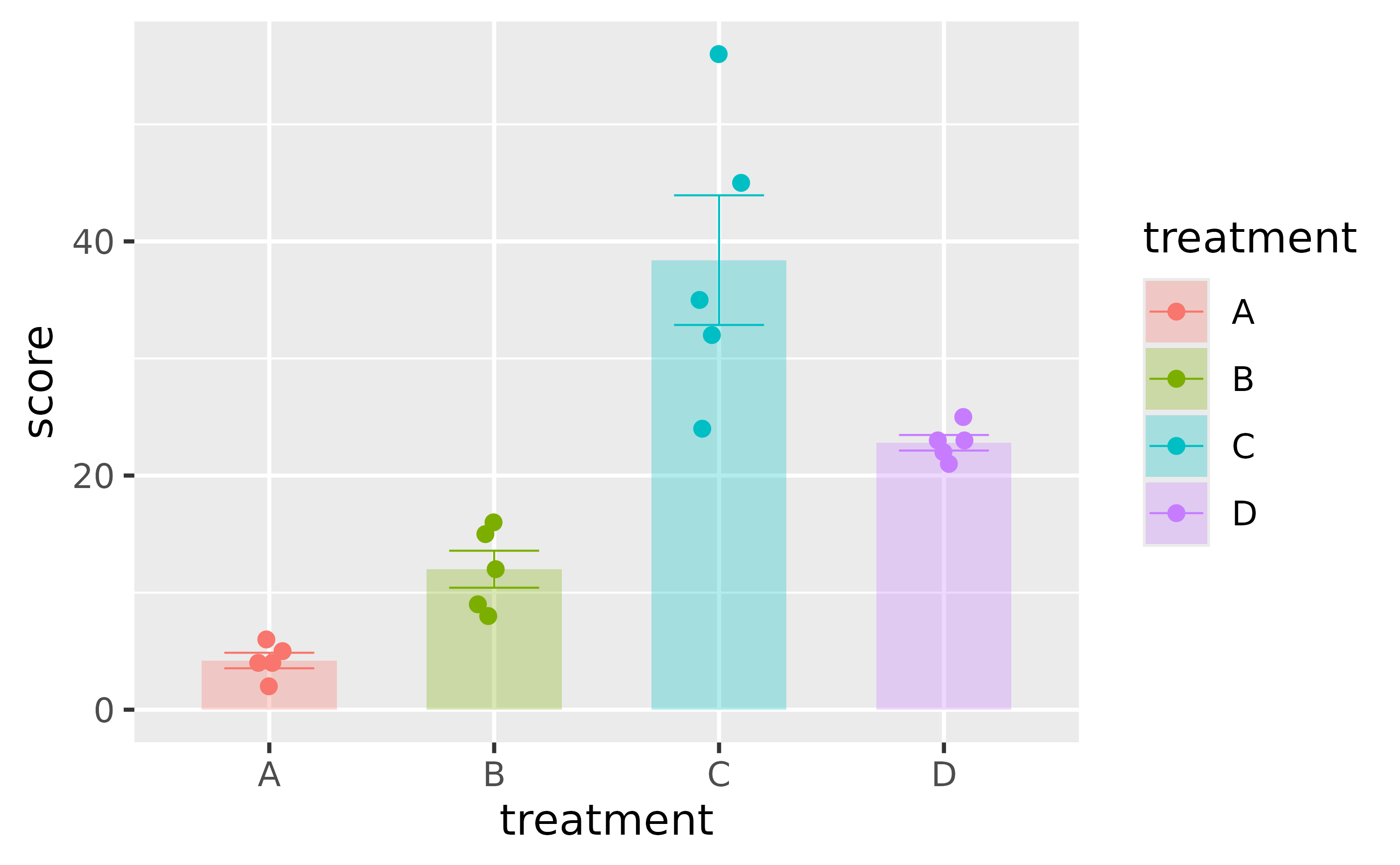
p$mapping
as_tp <-
p %>%
as_tidyplot() %>%
adjust_plot_area_padding(bottom = 0)
as_tp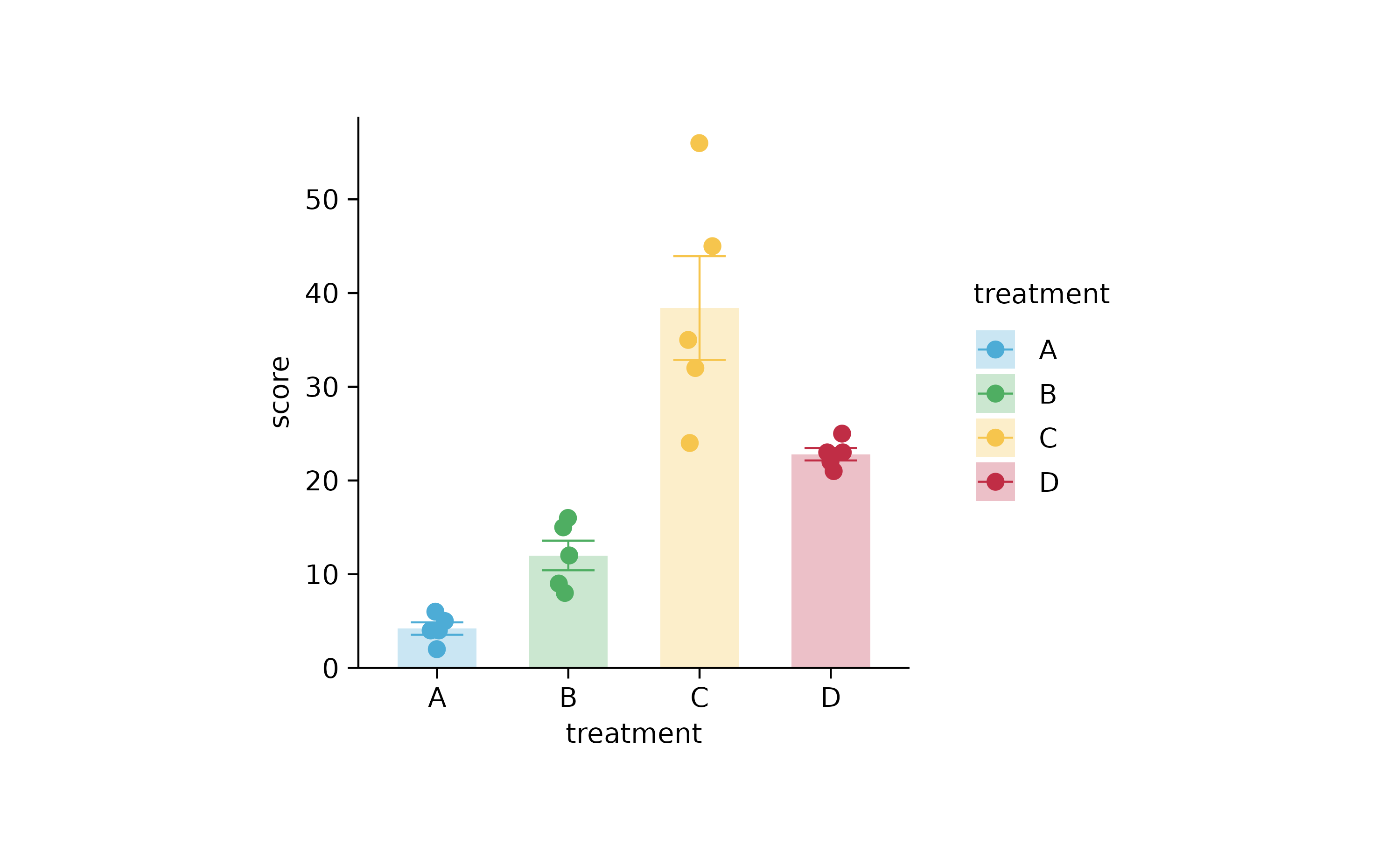
as_tp$tidyplot
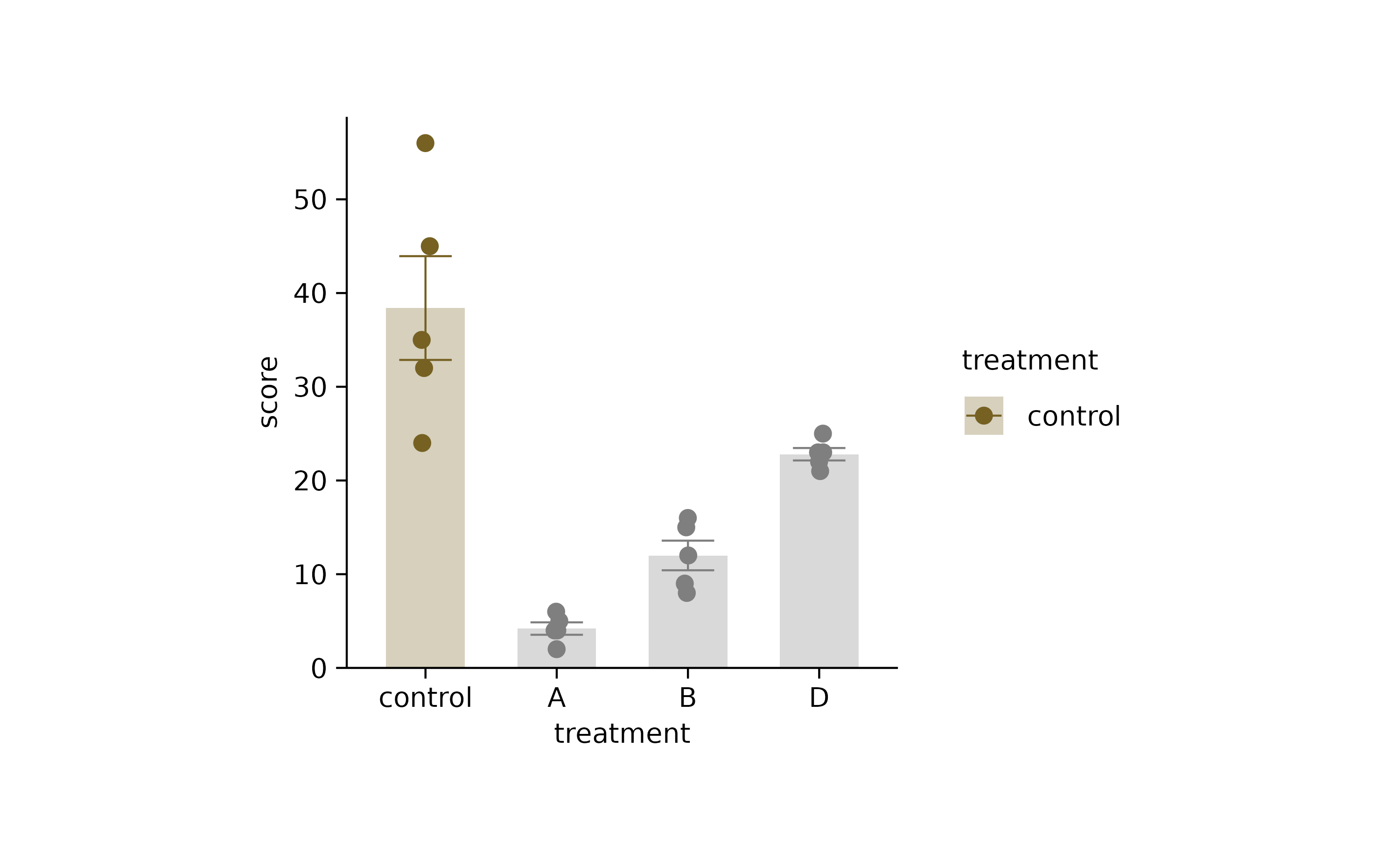
as_tp %>%
adjust_colors(new_colors = c("C" = "#766123")) %>%
reorder_x_axis_labels(c("C")) %>%
rename_x_axis_labels(c("C" = "control"))