The goal of bro_plot_heatmap() is to provide a tidyverse-style interface to the powerful heatmap package pheatmap by @raivokolde. Although heatmaps can be generated via ggplot2::geom_tile(), it is hard to reach the versatility and beauty of a genuine heatmap function like pheatmap::pheatmap().
Data requirements
bro_plot_heatmap() requires tidy data in long format, see tidyverse.
As an example we will use the gene expression data set bro_data_exprs. In the tidyverse lingo the columns of a data frame are called variables. One of these variables named expression contains the numeric values to be color-coded in the heatmap. Furthermore we will use the variables sample for heatmap columns and external_gene_name for heatmap rows.
bro_data_exprs
#> # A tibble: 800 x 12
#> ensembl_gene_id external_gene_na… Eip_vs_Hip.padj direction is_immune_gene
#> <chr> <chr> <dbl> <chr> <chr>
#> 1 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 2 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 3 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 4 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 5 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 6 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 7 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 8 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 9 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> 10 ENSMUSG00000033576 Apol6 3.79e-28 up no
#> # … with 790 more rows, and 7 more variables: count_type <chr>, group <chr>,
#> # replicate <chr>, expression <dbl>, sample <chr>, sample_type <chr>,
#> # condition <chr>
glimpse(bro_data_exprs)
#> Rows: 800
#> Columns: 12
#> $ ensembl_gene_id <chr> "ENSMUSG00000033576", "ENSMUSG00000033576", "ENSMUS…
#> $ external_gene_name <chr> "Apol6", "Apol6", "Apol6", "Apol6", "Apol6", "Apol6…
#> $ Eip_vs_Hip.padj <dbl> 3.793735e-28, 3.793735e-28, 3.793735e-28, 3.793735e…
#> $ direction <chr> "up", "up", "up", "up", "up", "up", "up", "up", "up…
#> $ is_immune_gene <chr> "no", "no", "no", "no", "no", "no", "no", "no", "no…
#> $ count_type <chr> "vsd", "vsd", "vsd", "vsd", "vsd", "vsd", "vsd", "v…
#> $ group <chr> "Hin", "Hin", "Hin", "Hin", "Hin", "Ein", "Ein", "E…
#> $ replicate <chr> "1", "2", "3", "4", "5", "1", "2", "3", "4", "5", "…
#> $ expression <dbl> 2.203755, 2.203755, 2.660558, 2.649534, 3.442740, 5…
#> $ sample <chr> "Hin_1", "Hin_2", "Hin_3", "Hin_4", "Hin_5", "Ein_1…
#> $ sample_type <chr> "input", "input", "input", "input", "input", "input…
#> $ condition <chr> "healthy", "healthy", "healthy", "healthy", "health…Basic usage
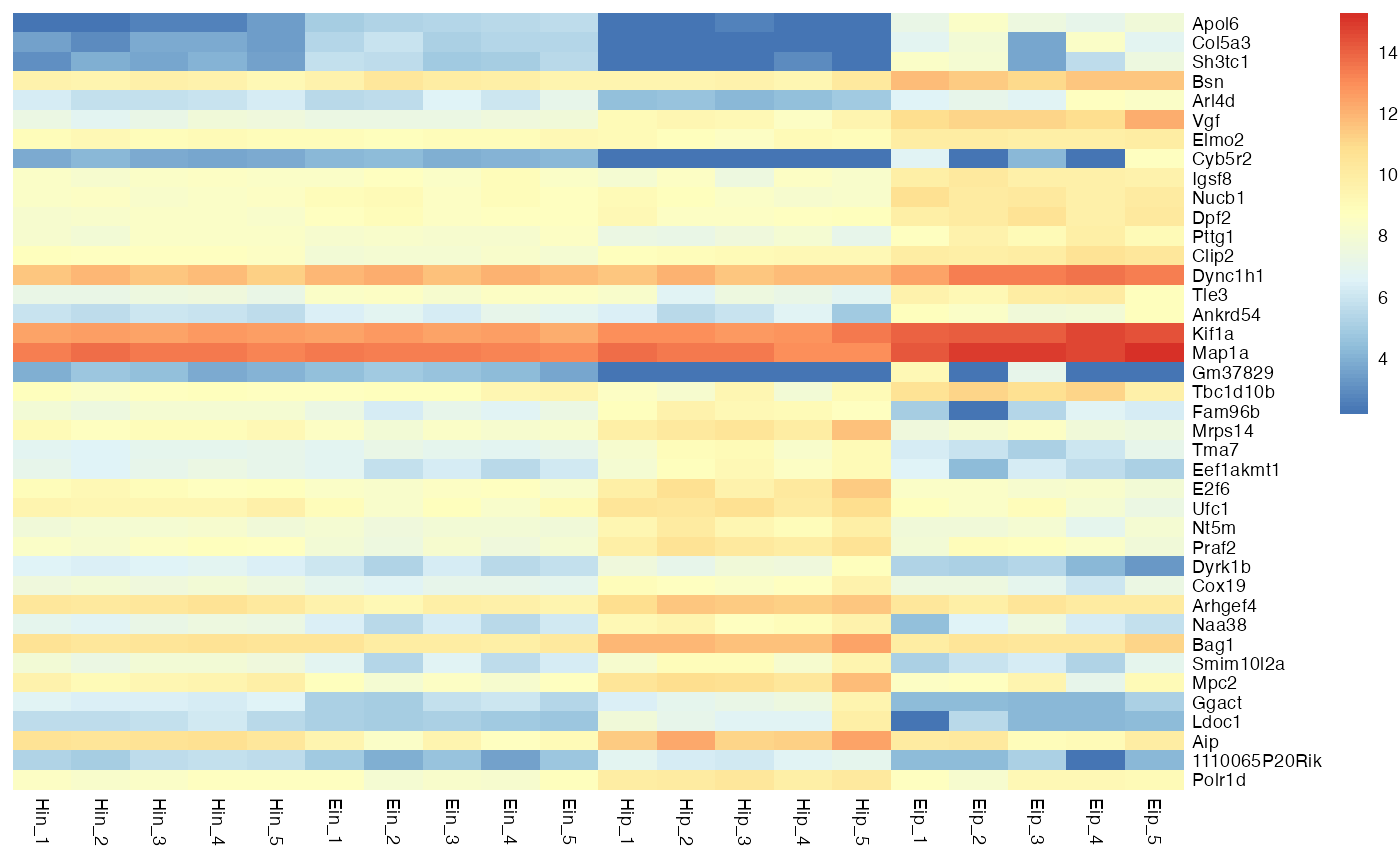
The basic layout of the heatmap relies on the parameters rows, columns and values. You can think of them like aesthetics in ggplot2::ggplot(), similar to something like aes(x = columns, y = rows, fill = values).
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression
)
Features
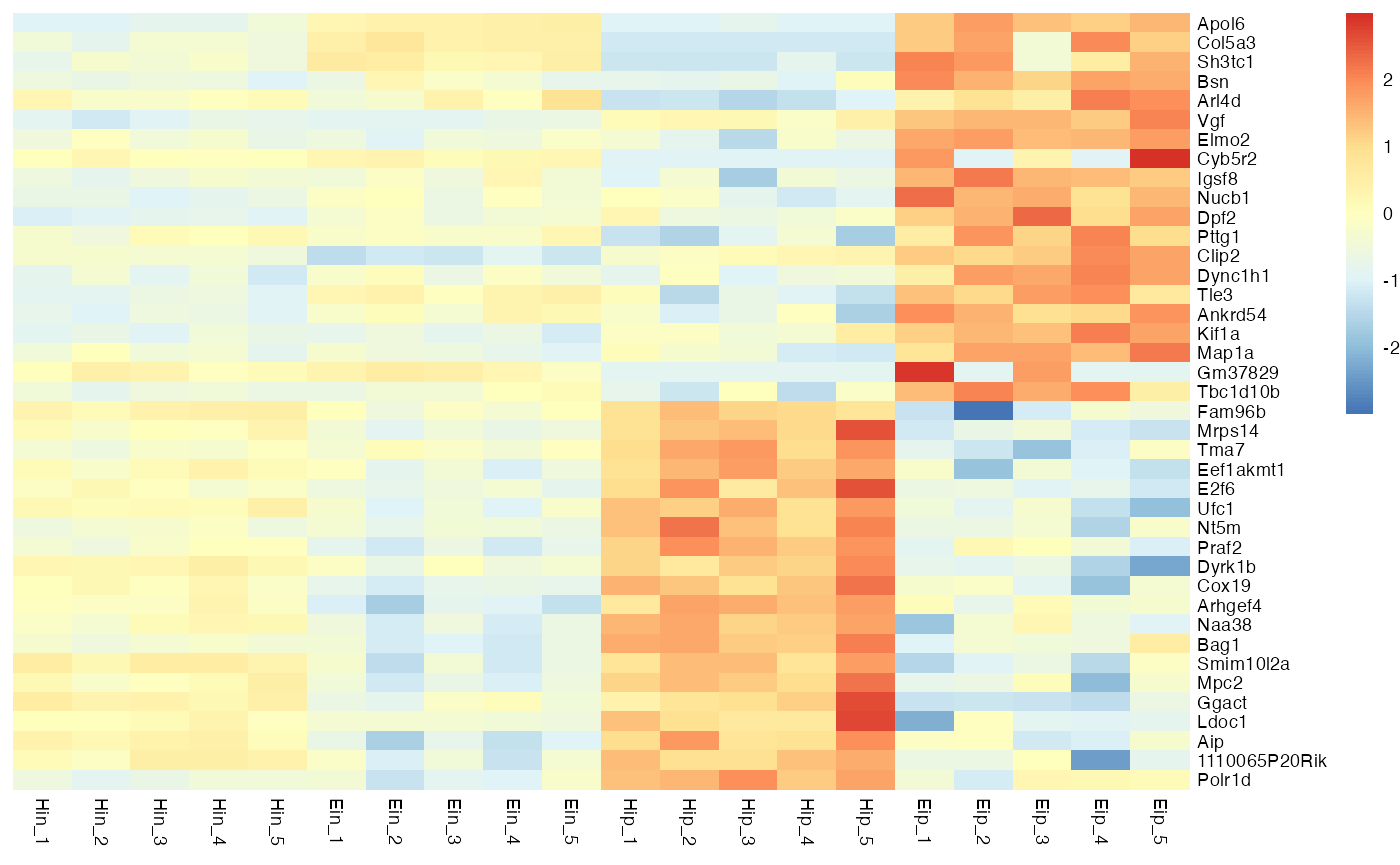
Data scaling
With the parameter scale you can activate data scaling for "row" or "column". By default data scaling is turned off scale = "none".
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
scale = "row"
)
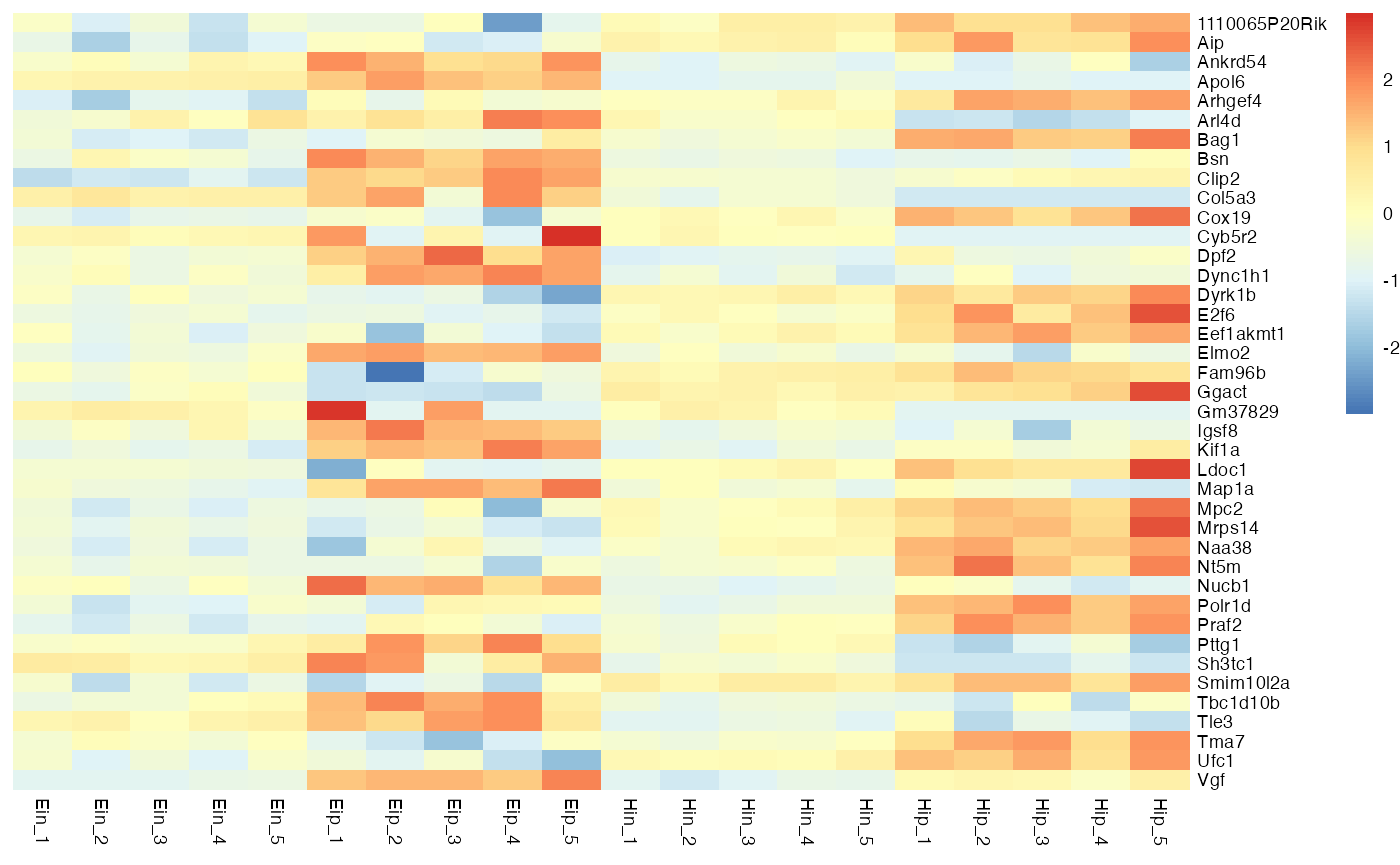
Ordering
Rows and columns in the heatmap will appear in the same order as in the tidy data frame used as input. For example, to order rows and columns alphabetically, just use the dplyr::arrange().
bro_data_exprs %>%
arrange(external_gene_name, sample) %>%
bro_plot_heatmap(rows = external_gene_name,
columns = sample,
values = expression,
scale = "row"
)
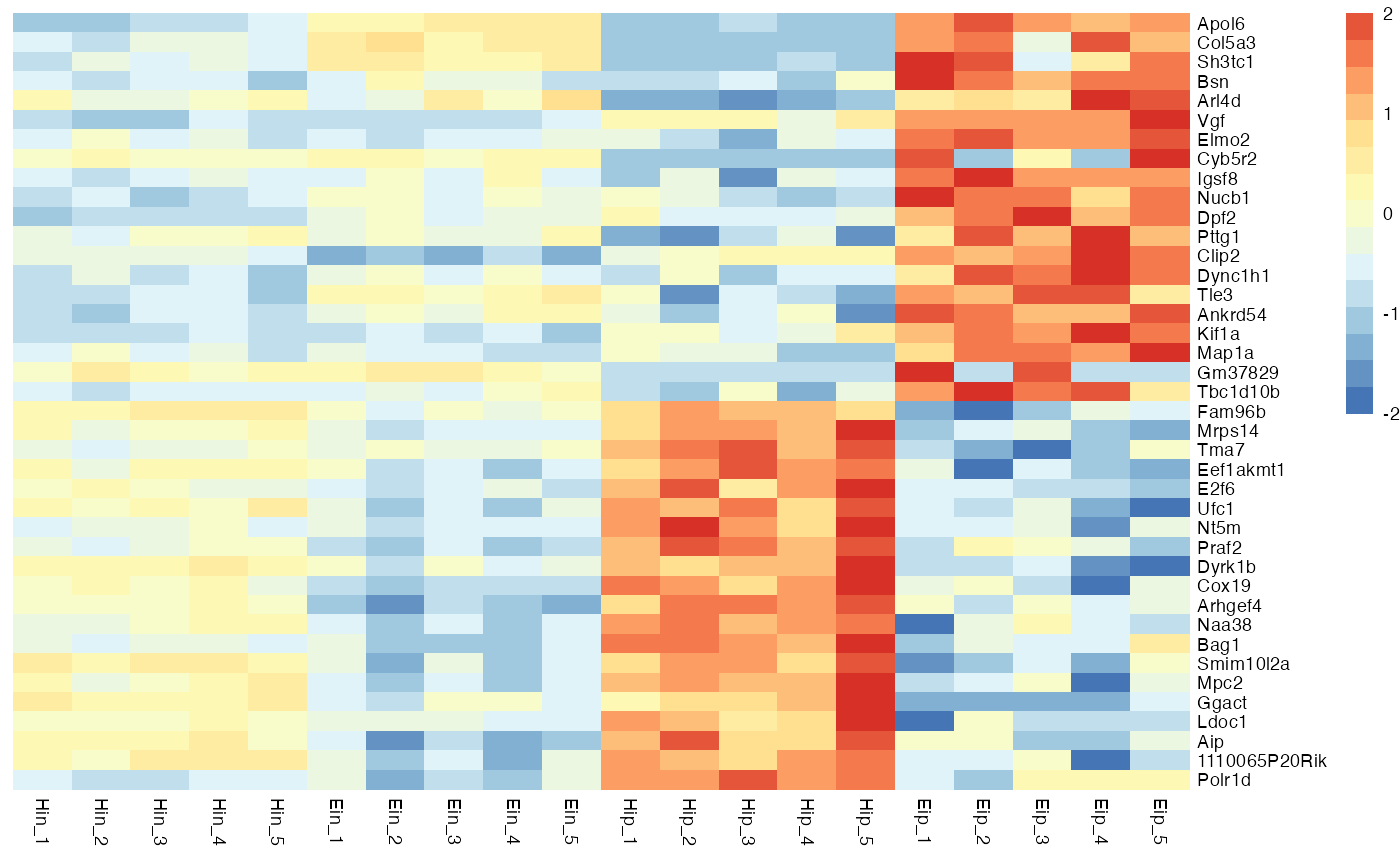
Color legend
You can customize the number of colors color_scale_n and also the minimum and maximum values of the color legend, color_scale_min and color_scale_max.
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
scale = "row",
color_scale_n = 16,
color_scale_min = -2,
color_scale_max = 2
)
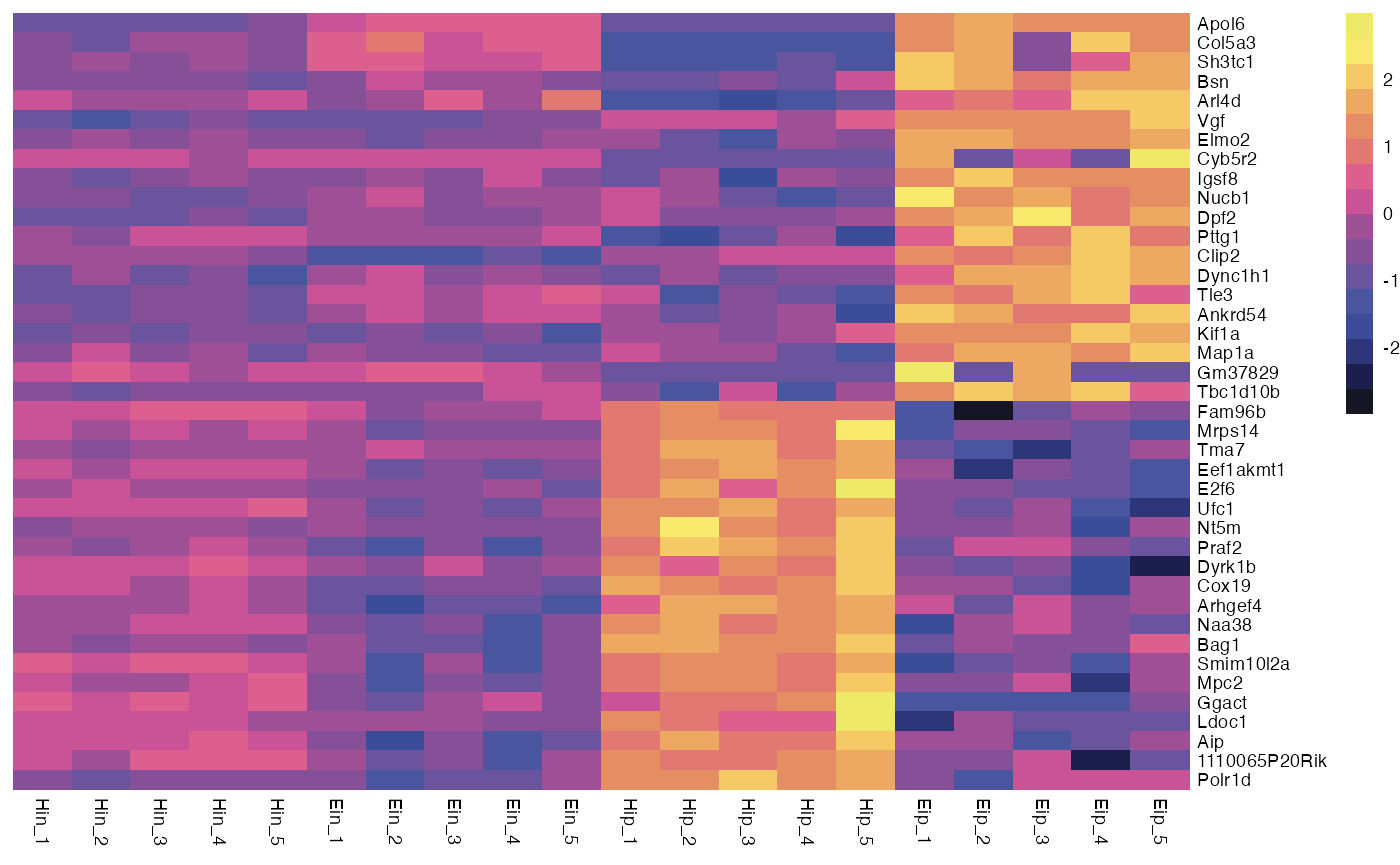
Of course, you can also replace the color legend altogether.
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
scale = "row",
color = c("#161523","#1C1E4C","#2D3679","#3B4D98","#49569F","#69549D",
"#855097","#9E4F96","#CA5296","#DC5F8E","#E17872","#E58D63",
"#EDA962","#F5CA66","#F9E96C","#EEE969")
)
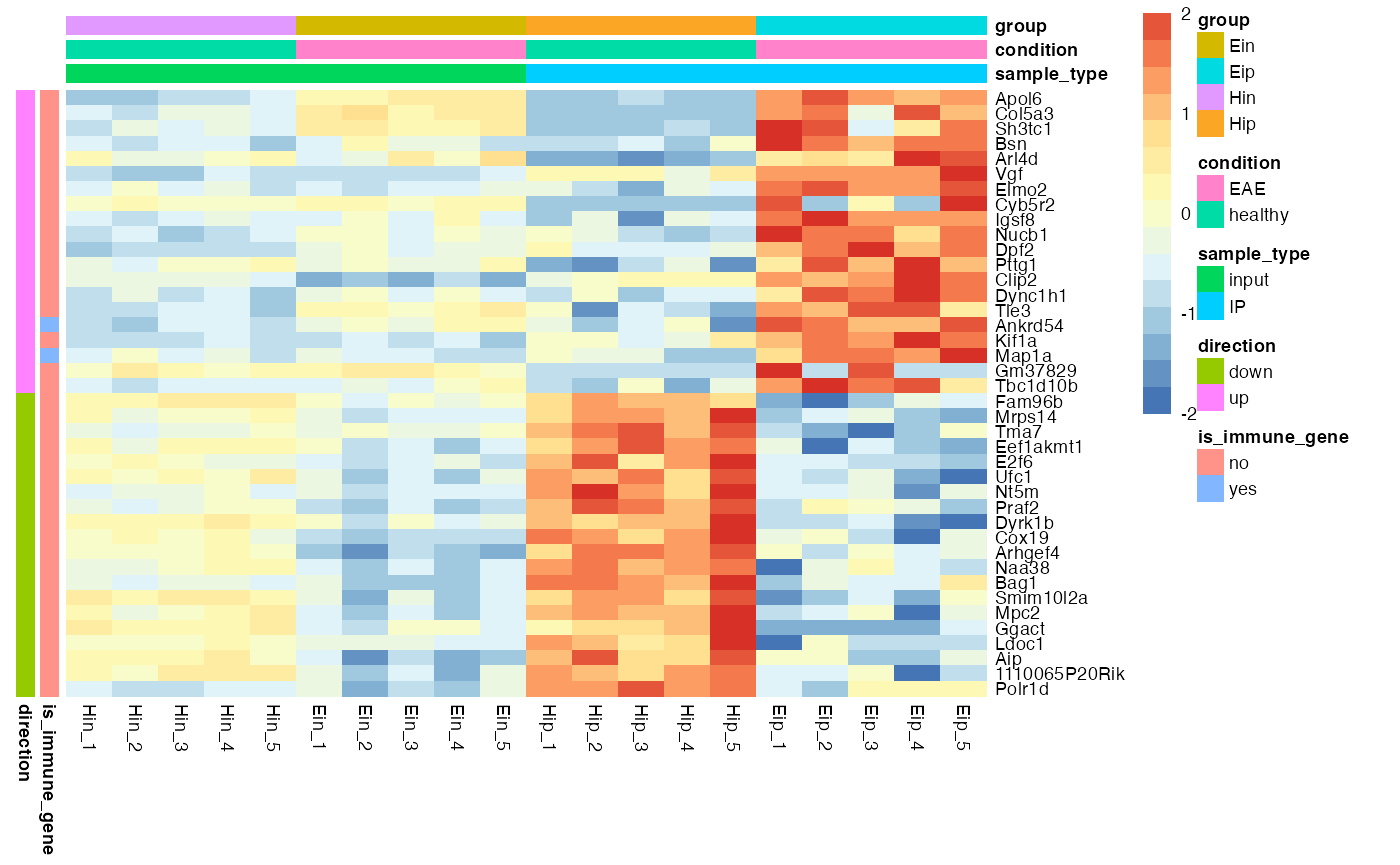
Annotations
Annotations can be added for both rows and columns via ann_row and ann_col, respectively. Just specify the corresponding variables in the tidy data frame. If you want more then one variable for annotation just combine them by c(var1, var2, var3).
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
ann_col = c(sample_type, condition, group),
ann_row = c(is_immune_gene, direction),
scale = "row",
color_scale_n = 16,
color_scale_min = -2,
color_scale_max = 2
)
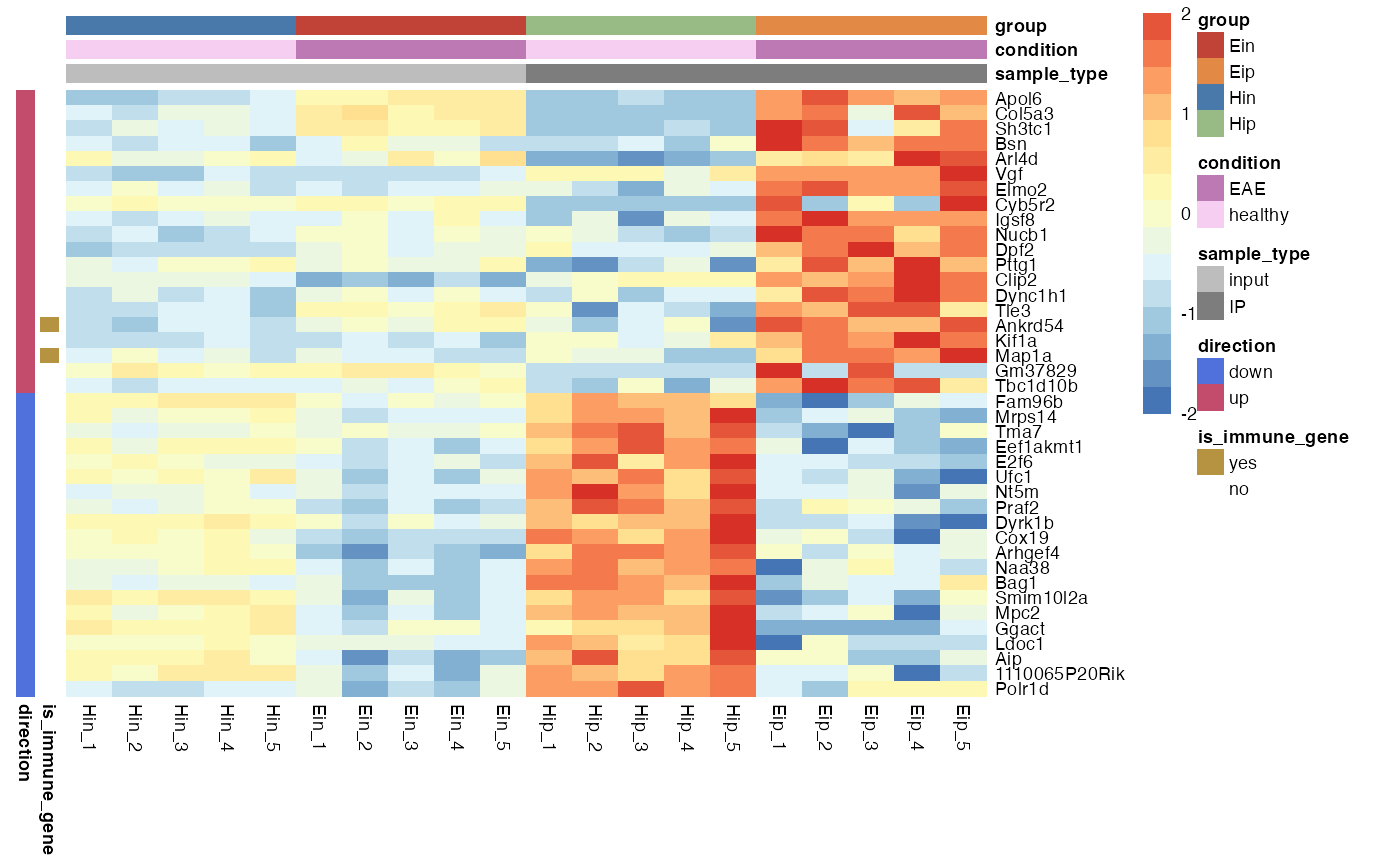
Customize annotations colors
You can provide a list of named vectors to take control over the annotations colors ann_colors.
ann_colors <- list(
condition = c(EAE = "#BD79B4", healthy = "#F5CEF2"),
group = c(Ein = "#C14236", Eip = "#E28946", Hin = "#4978AB", Hip = "#98BB85"),
sample_type = c(input = "#BDBDBD", IP = "#7D7D7D"),
direction = c(down = "#5071DC", up = "#C34B6B"),
is_immune_gene = c(yes = "#B69340", no = "#FFFFFF")
)
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
ann_col = c(sample_type, condition, group),
ann_row = c(is_immune_gene, direction),
scale = "row",
color_scale_n = 16,
color_scale_min = -2,
color_scale_max = 2,
ann_colors = ann_colors
)
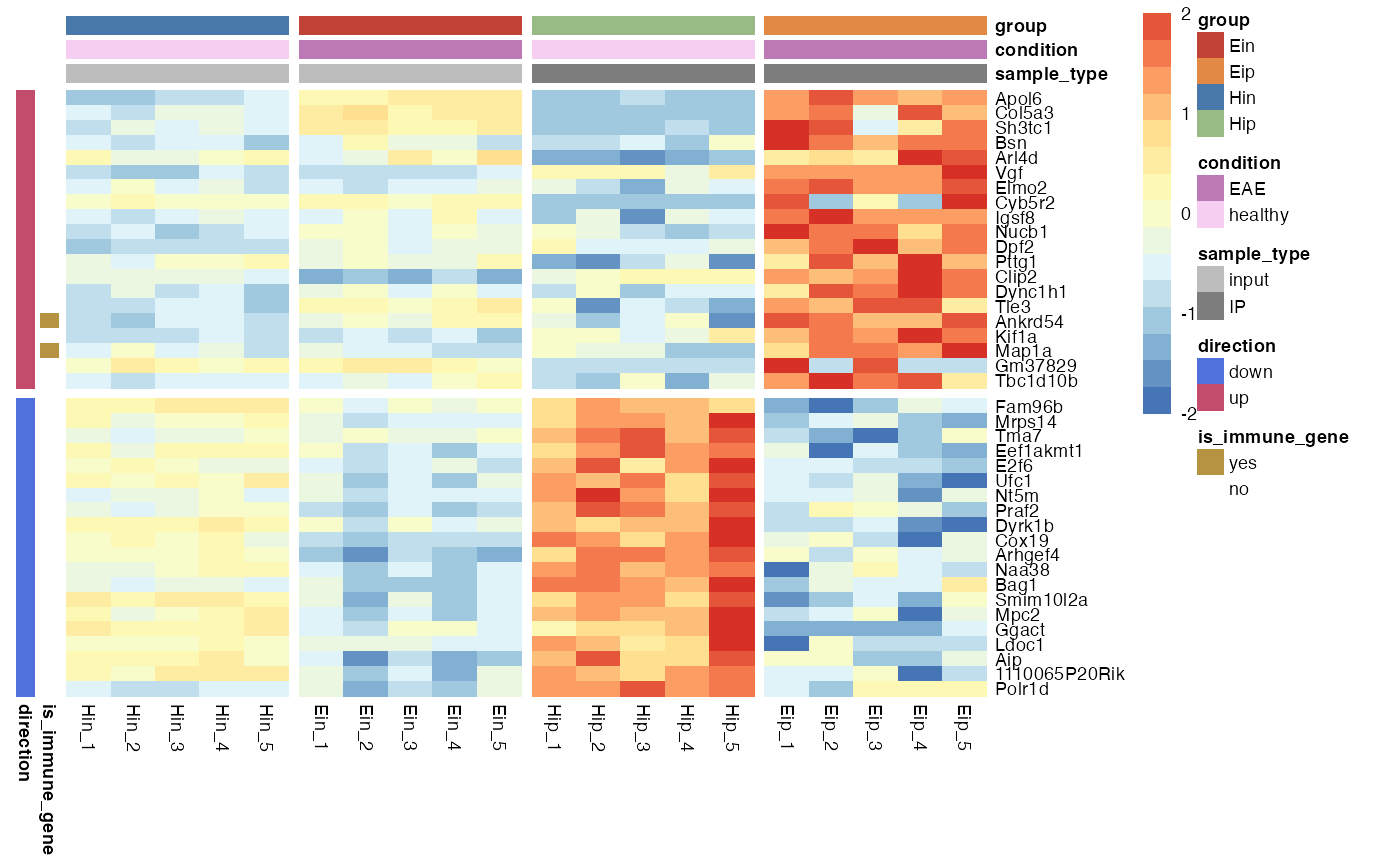
Gaps
Gaps can be added by specifying data frame variables that should be used to generate the gaps. Only one variable can be chosen for gaps_row and one for gaps_col.
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
ann_col = c(sample_type, condition, group),
ann_row = c(is_immune_gene, direction),
scale = "row",
color_scale_n = 16,
color_scale_min = -2,
color_scale_max = 2,
ann_colors = ann_colors,
gaps_row = direction,
gaps_col = group
)
Fix cell dimensions
You can fix the cell dimensions via the cellwidth and cellheight parameters.
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
ann_col = c(sample_type, condition, group),
ann_row = c(is_immune_gene, direction),
scale = "row",
color_scale_n = 16,
color_scale_min = -2,
color_scale_max = 2,
ann_colors = ann_colors,
gaps_row = direction,
gaps_col = group,
cellwidth = 7,
cellheight = 7
)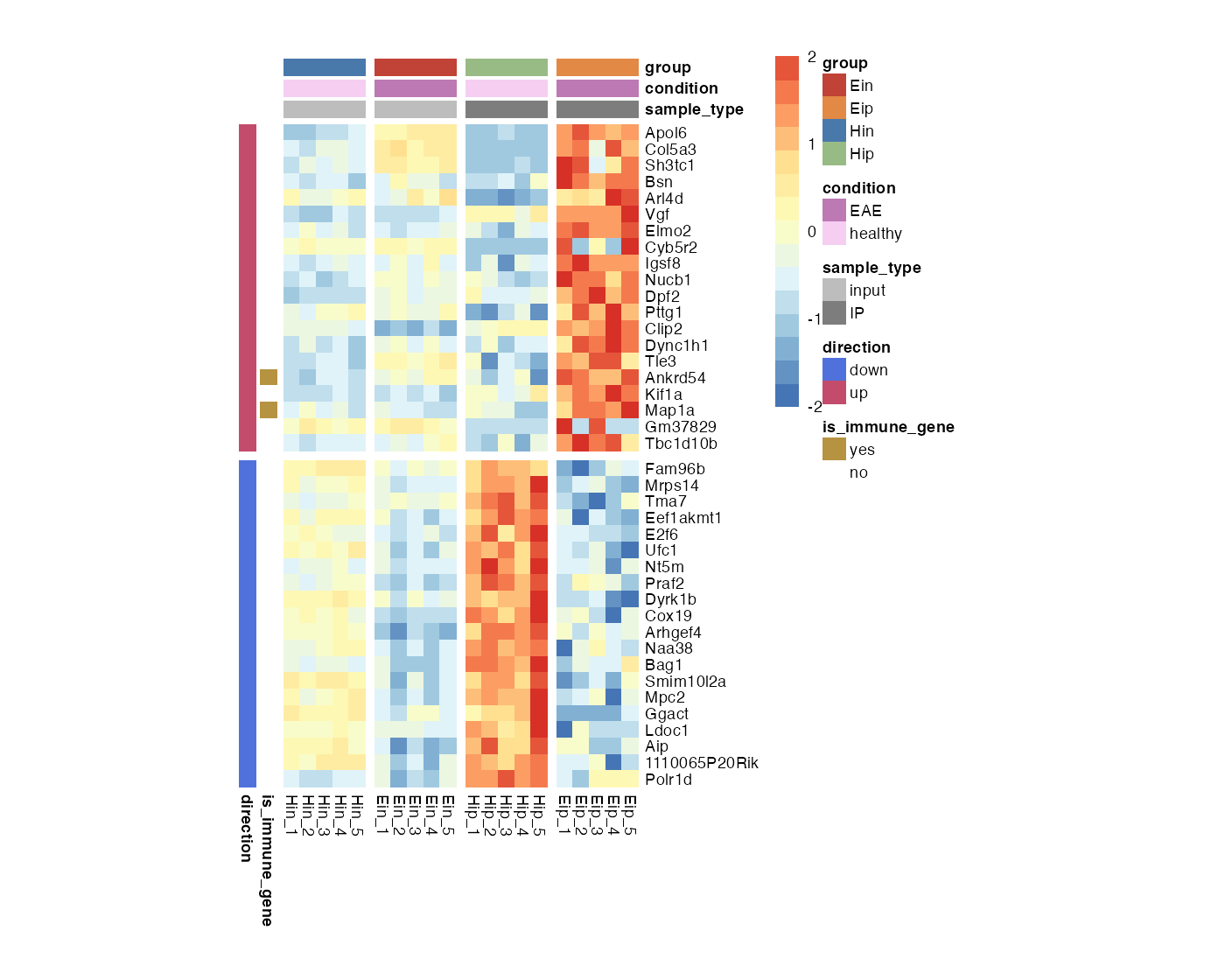
Write to file
You can use the parameter filename to write the heatmap to file.
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
filename = "my_heatmap.pdf"
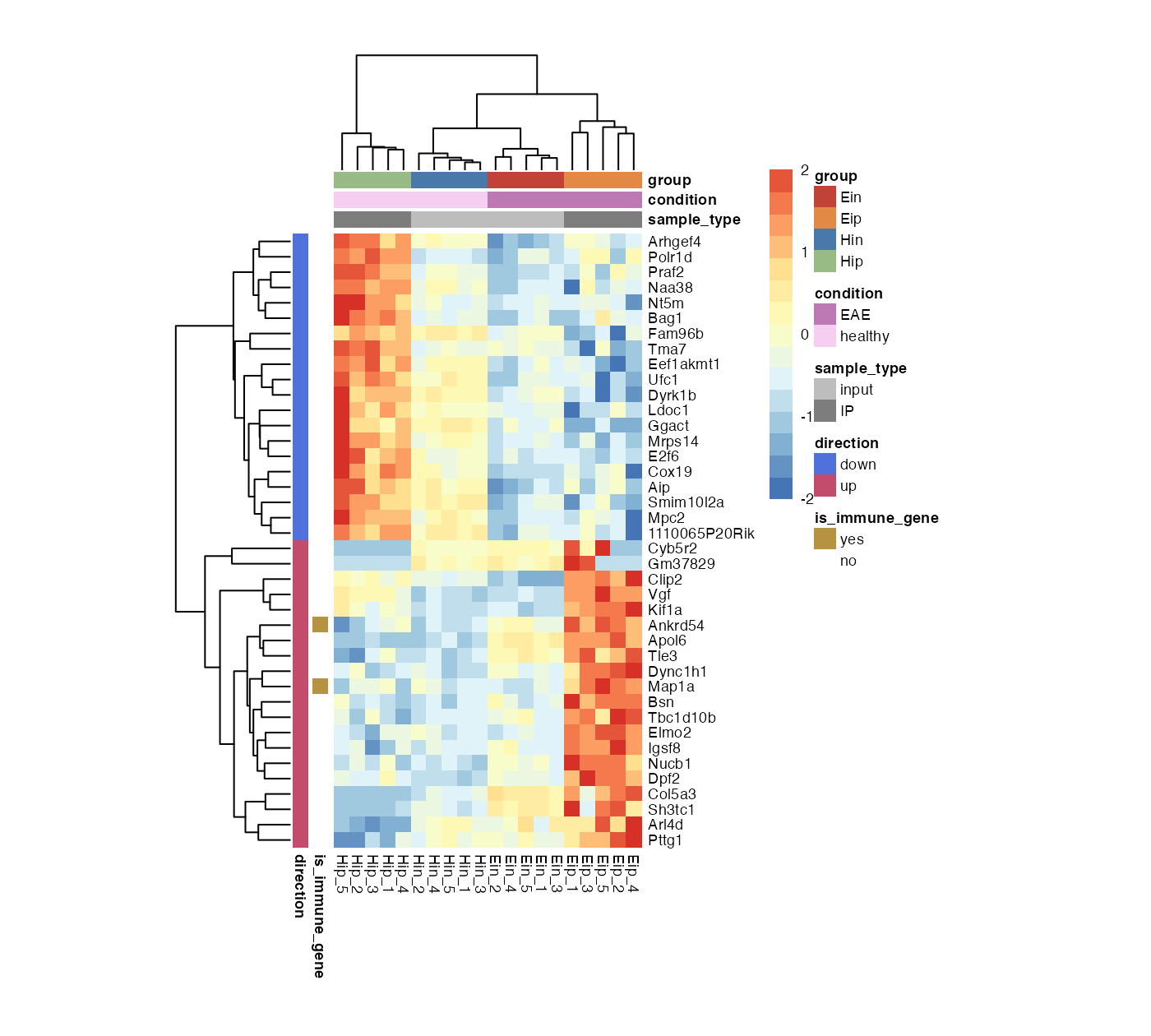
)More features
There are even more features like clustering and dendrograms implemented in pheatmap::pheatmap(). All parameters that are not directly evaluated by bro_plot_heatmap() will be automatically passed to pheatmap::pheatmap().
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
ann_col = c(sample_type, condition, group),
ann_row = c(is_immune_gene, direction),
scale = "row",
color_scale_n = 16,
color_scale_min = -2,
color_scale_max = 2,
ann_colors = ann_colors,
cellwidth = 7,
cellheight = 7,
cluster_rows = TRUE,
cluster_cols = TRUE
)
What’s under the hood?
bro_plot_heatmap() takes the tidy data frame and the additional arguments and does the data wrangling needed to satisfy pheatmap::pheatmap(). By setting return_data = TRUE you can get a glimpse at the result of the data wrangling.
bro_plot_heatmap(bro_data_exprs,
rows = external_gene_name,
columns = sample,
values = expression,
ann_col = c(sample_type, condition, group),
ann_row = c(is_immune_gene, direction),
gaps_row = direction,
gaps_col = group,
return_data = TRUE
)
#> $m
#> Hin_1 Hin_2 Hin_3 Hin_4 Hin_5 Ein_1
#> Apol6 2.203755 2.203755 2.660558 2.649534 3.442740 5.030997
#> Col5a3 3.623289 2.950514 3.883794 3.821838 3.486696 5.347029
#> Sh3tc1 3.094325 3.945669 3.721781 4.071979 3.569361 5.758298
#> Bsn 9.594692 9.481155 9.663027 9.621492 9.254889 9.549501
#> Arl4d 6.379306 5.795141 5.767258 5.991696 6.256381 5.513601
#> Vgf 7.386505 6.861794 7.266003 7.738055 7.575901 7.385140
#> Elmo2 8.964594 9.191505 8.996498 9.091161 8.877599 8.940428
#> Cyb5r2 3.846136 4.181117 3.778346 3.687823 3.817430 4.242324
#> Igsf8 8.357730 8.182801 8.386692 8.557224 8.453666 8.401367
#> Nucb1 8.472584 8.486825 8.245469 8.378464 8.521781 8.934631
#> Dpf2 8.209568 8.291422 8.374594 8.442385 8.289120 8.723934
#> Pttg1 8.137118 7.937426 8.404651 8.352276 8.469586 8.156889
#> Clip2 8.754678 8.733512 8.694684 8.715489 8.554136 7.846969
#> Dync1h1 11.602393 11.942333 11.551018 11.851763 11.346086 12.000915
#> Tle3 7.289507 7.292232 7.546951 7.607970 7.198383 8.416004
#> Ankrd54 5.867274 5.619651 6.048261 5.986635 5.665235 6.463674
#> Kif1a 12.437160 12.581432 12.410019 12.770907 12.579293 12.524093
#> Map1a 13.417764 13.772932 13.446422 13.530706 13.201875 13.563318
#> Gm37829 3.911792 4.704230 4.474480 3.796259 4.124560 4.462020
#> Tbc1d10b 8.827458 8.456042 8.726761 8.808308 8.656032 8.639146
#> Fam96b 7.835157 7.517774 7.957167 8.045903 8.081315 7.301972
#> Mrps14 9.088216 8.681238 8.922262 8.876584 9.258166 8.500837
#> Tma7 6.811239 6.566974 6.949656 6.908019 7.116900 6.844333
#> Eef1akmt1 7.099069 6.641980 7.059650 7.361731 7.093585 6.844333
#> E2f6 8.896741 9.150440 8.964573 8.654886 8.829067 8.465089
#> Ufc1 9.416171 9.296585 9.387115 9.321553 9.659070 8.983176
#> Nt5m 7.804291 7.995676 8.048851 8.187596 7.817257 7.979595
#> Praf2 8.381444 8.195282 8.537184 8.777298 8.709678 7.945404
#> Dyrk1b 6.547531 6.467346 6.543533 6.825701 6.429647 6.044776
#> Cox19 7.678025 7.837444 7.647610 7.914175 7.539717 7.035425
#> Arhgef4 10.341459 10.299102 10.306279 10.569326 10.301059 9.643109
#> Naa38 6.916126 6.660135 7.287685 7.481177 7.418356 6.397601
#> Bag1 10.574157 10.398226 10.500136 10.594572 10.454331 10.484227
#> Smim10l2a 7.894974 7.404043 7.899221 7.873827 7.571691 6.825633
#> Mpc2 9.548895 9.108171 9.299191 9.511867 9.882313 8.854007
#> Ggact 6.739533 6.404070 6.492079 6.258252 6.592570 5.137649
#> Ldoc1 5.691651 5.692469 5.816111 6.162898 5.531716 5.095964
#> Aip 10.641707 10.466733 10.643964 10.766897 10.318114 9.478571
#> 1110065P20Rik 5.302977 5.024172 5.703733 5.750027 5.616628 4.939347
#> Polr1d 8.553674 8.320197 8.445705 8.649320 8.645059 8.669130
#> Ein_2 Ein_3 Ein_4 Ein_5 Hip_1 Hip_2
#> Apol6 5.314412 5.372403 5.536960 5.645714 2.203755 2.203755
#> Col5a3 5.970917 5.200783 5.380863 5.414576 2.203755 2.203755
#> Sh3tc1 5.628464 4.943715 5.004403 5.505737 2.203755 2.203755
#> Bsn 10.336688 9.978072 9.823801 9.465946 9.427566 9.401769
#> Arl4d 5.697178 6.555629 6.031178 7.073764 4.493476 4.561038
#> Vgf 7.394351 7.393358 7.676219 7.781865 9.049624 9.300449
#> Elmo2 8.777253 8.986457 8.935158 9.130373 9.052209 8.822092
#> Cyb5r2 4.330928 3.906655 4.122089 4.180508 2.203755 2.203755
#> Igsf8 8.692153 8.365600 8.955830 8.445110 8.056258 8.517292
#> Nucb1 9.042499 8.533012 8.976209 8.717655 9.020879 8.859275
#> Dpf2 8.901595 8.525343 8.685763 8.714508 9.131351 8.539024
#> Pttg1 8.262570 8.153131 8.188274 8.504625 7.394518 7.203414
#> Clip2 8.004669 7.980647 8.256524 7.994040 8.739036 8.891307
#> Dync1h1 12.203191 11.700997 12.095691 11.835714 11.587370 12.117493
#> Tle3 8.538112 8.166341 8.529046 8.603207 8.275674 6.664542
#> Ankrd54 6.806487 6.350472 7.038793 6.907131 6.425222 5.510484
#> Kif1a 12.726740 12.468208 12.601176 12.239755 13.040172 13.008885
#> Map1a 13.414518 13.374582 13.211265 13.125640 13.819813 13.550660
#> Gm37829 4.909113 4.627398 4.316214 3.711095 2.203755 2.203755
#> Tbc1d10b 8.861327 8.849873 9.273811 9.404788 8.528672 8.086715
#> Fam96b 6.356636 7.066705 6.689155 7.317450 8.756578 9.641596
#> Mrps14 7.942847 8.427253 8.188274 8.342835 9.832043 10.285114
#> Tma7 7.254255 7.016478 6.784851 7.150363 8.193745 8.906015
#> Eef1akmt1 5.794391 6.338816 5.536960 6.228244 8.030265 8.767910
#> E2f6 8.290413 8.496872 8.685763 8.219487 9.891710 10.713109
#> Ufc1 8.333873 8.823020 8.300294 9.017444 10.487858 10.344933
#> Nt5m 7.665054 7.938917 7.891876 7.775837 9.325486 10.060336
#> Praf2 7.549944 8.088675 7.608173 8.004387 9.811593 10.681098
#> Dyrk1b 5.269735 6.254461 5.536960 5.797272 7.627413 7.060328
#> Cox19 6.759376 7.038219 7.089617 7.033880 8.961613 8.730623
#> Arhgef4 9.170855 9.791434 9.706727 9.408687 10.840547 11.551055
#> Naa38 5.556250 6.373502 5.536960 6.210416 9.131351 9.457031
#> Bag1 9.941392 10.009557 9.893316 10.284755 11.955751 12.001475
#> Smim10l2a 5.357711 6.662247 5.608986 6.363402 8.146133 8.967404
#> Mpc2 8.011377 8.600253 8.140910 8.714508 10.555920 10.918746
#> Ggact 4.966731 5.839785 6.082447 5.382831 6.480152 7.029922
#> Ldoc1 5.075229 5.147621 4.891888 4.792294 7.698471 7.090106
#> Aip 8.360385 9.399647 8.734433 9.125643 11.376675 12.298776
#> 1110065P20Rik 3.904594 4.587228 3.606475 4.742274 6.839297 6.254633
#> Polr1d 8.024701 8.301193 8.211385 8.803094 9.974441 10.069697
#> Hip_3 Hip_4 Hip_5 Eip_1 Eip_2 Eip_3
#> Apol6 2.619196 2.203755 2.203755 7.207838 8.418188 7.505587
#> Col5a3 2.203755 2.203755 2.203755 6.900971 7.825300 3.719267
#> Sh3tc1 2.203755 2.932447 2.203755 8.434457 8.051111 3.719267
#> Bsn 9.520877 9.264847 10.194225 11.871814 11.444950 11.083346
#> Arl4d 4.205448 4.438281 4.835291 6.552481 7.099169 6.695912
#> Vgf 9.204563 8.585649 9.514597 10.931662 11.184191 11.176536
#> Elmo2 8.525198 9.101038 8.908268 9.970747 10.031208 9.857982
#> Cyb5r2 2.203755 2.203755 2.203755 6.728107 2.203755 4.266641
#> Igsf8 7.506240 8.486750 8.279999 9.808985 10.289182 9.777884
#> Nucb1 8.391101 8.083832 8.309137 10.775210 10.149783 10.243176
#> Dpf2 8.502422 8.623033 8.841703 9.802469 10.066092 10.665406
#> Pttg1 7.689547 8.076149 7.097184 8.709842 9.649422 9.101513
#> Clip2 9.086007 9.180797 9.213187 9.966868 9.802079 9.933866
#> Dync1h1 11.481329 11.809939 11.841209 12.494681 13.397535 13.314656
#> Tle3 7.493971 7.238095 6.794339 9.548462 9.224495 9.958299
#> Ankrd54 5.953090 6.674710 4.889054 8.830037 8.418188 7.749207
#> Kif1a 12.765709 12.826279 13.523318 13.965006 14.222354 14.103765
#> Map1a 13.495391 12.977465 12.929212 14.300342 14.931126 14.920673
#> Gm37829 2.203755 2.203755 2.203755 9.246378 2.203755 7.156942
#> Tbc1d10b 9.274336 7.894408 9.084301 10.594685 11.215603 10.784057
#> Fam96b 9.161602 9.031911 8.667046 5.036369 2.203755 5.396467
#> Mrps14 10.446139 10.017238 11.681903 7.627301 8.184126 8.491745
#> Tma7 9.061288 8.231774 9.120360 6.303794 5.908813 5.188813
#> Eef1akmt1 9.211133 8.529567 9.025541 6.593407 4.392886 6.345960
#> E2f6 9.538148 10.242673 11.423428 8.411804 8.471214 8.139536
#> Ufc1 10.697065 10.061670 10.952405 8.825760 8.418188 8.995318
#> Nt5m 9.326266 8.935445 9.889655 7.749807 7.741421 8.020793
#> Praf2 10.220552 9.916178 10.587568 7.854334 8.953492 8.737433
#> Dyrk1b 7.726716 7.583559 8.789252 5.259213 5.096047 5.396467
#> Cox19 8.382811 8.730516 9.530145 7.438400 7.557424 6.911790
#> Arhgef4 11.459819 11.299810 11.583997 10.404160 9.822626 10.452586
#> Naa38 8.684764 8.869578 9.539394 4.530752 6.623996 7.505587
#> Bag1 11.721443 11.641706 12.408304 9.989989 10.520580 10.411887
#> Smim10l2a 8.992131 8.145087 9.397410 5.152236 5.908813 6.345960
#> Mpc2 10.697065 10.431086 11.836423 8.511054 8.665741 9.427408
#> Ggact 7.197444 7.499866 9.511467 4.342894 4.392886 4.266641
#> Ldoc1 6.651752 6.691598 9.855504 2.203755 5.560057 4.266641
#> Aip 11.120444 11.242921 12.433740 10.157616 10.213406 8.914373
#> 1110065P20Rik 6.208129 6.760435 7.016961 4.342894 4.392886 5.188813
#> Polr1d 10.414774 9.896324 10.241586 8.667443 8.119152 9.158861
#> Eip_4 Eip_5
#> Apol6 7.116522 7.692020
#> Col5a3 8.444539 6.795068
#> Sh3tc1 5.605475 7.439714
#> Bsn 11.585093 11.489941
#> Arl4d 8.637639 8.449852
#> Vgf 10.864963 12.160344
#> Elmo2 9.872046 10.027214
#> Cyb5r2 2.203755 8.632506
#> Igsf8 9.726716 9.615808
#> Nucb1 9.657107 10.140765
#> Dpf2 9.674827 10.185692
#> Pttg1 9.793121 9.031047
#> Clip2 10.594067 10.339874
#> Dync1h1 13.595866 13.354095
#> Tle3 10.125012 8.835462
#> Ankrd54 7.836834 8.773737
#> Kif1a 14.682213 14.385192
#> Map1a 14.732255 15.269623
#> Gm37829 2.203755 2.203755
#> Tbc1d10b 11.092944 9.728544
#> Fam96b 6.765439 6.373782
#> Mrps14 7.705013 7.481524
#> Tma7 6.103697 7.052702
#> Eef1akmt1 5.605475 5.145525
#> E2f6 8.268975 7.898976
#> Ufc1 8.014395 7.418346
#> Nt5m 7.008741 8.073099
#> Praf2 8.359427 7.718482
#> Dyrk1b 4.170949 3.253924
#> Cox19 6.103697 7.385686
#> Arhgef4 10.072506 10.171403
#> Naa38 6.299870 5.807935
#> Bag1 10.317786 11.193410
#> Smim10l2a 5.270378 6.996101
#> Mpc2 7.116522 9.034560
#> Ggact 4.170949 5.197424
#> Ldoc1 4.170949 4.389168
#> Aip 9.044393 9.930596
#> 1110065P20Rik 2.203755 4.190115
#> Polr1d 9.149524 9.089636
#>
#> $ann_row
#> is_immune_gene direction
#> Apol6 no up
#> Col5a3 no up
#> Sh3tc1 no up
#> Bsn no up
#> Arl4d no up
#> Vgf no up
#> Elmo2 no up
#> Cyb5r2 no up
#> Igsf8 no up
#> Nucb1 no up
#> Dpf2 no up
#> Pttg1 no up
#> Clip2 no up
#> Dync1h1 no up
#> Tle3 no up
#> Ankrd54 yes up
#> Kif1a no up
#> Map1a yes up
#> Gm37829 no up
#> Tbc1d10b no up
#> Fam96b no down
#> Mrps14 no down
#> Tma7 no down
#> Eef1akmt1 no down
#> E2f6 no down
#> Ufc1 no down
#> Nt5m no down
#> Praf2 no down
#> Dyrk1b no down
#> Cox19 no down
#> Arhgef4 no down
#> Naa38 no down
#> Bag1 no down
#> Smim10l2a no down
#> Mpc2 no down
#> Ggact no down
#> Ldoc1 no down
#> Aip no down
#> 1110065P20Rik no down
#> Polr1d no down
#>
#> $ann_col
#> sample_type condition group
#> Hin_1 input healthy Hin
#> Hin_2 input healthy Hin
#> Hin_3 input healthy Hin
#> Hin_4 input healthy Hin
#> Hin_5 input healthy Hin
#> Ein_1 input EAE Ein
#> Ein_2 input EAE Ein
#> Ein_3 input EAE Ein
#> Ein_4 input EAE Ein
#> Ein_5 input EAE Ein
#> Hip_1 IP healthy Hip
#> Hip_2 IP healthy Hip
#> Hip_3 IP healthy Hip
#> Hip_4 IP healthy Hip
#> Hip_5 IP healthy Hip
#> Eip_1 IP EAE Eip
#> Eip_2 IP EAE Eip
#> Eip_3 IP EAE Eip
#> Eip_4 IP EAE Eip
#> Eip_5 IP EAE Eip
#>
#> $gaps_row
#> [1] 20
#>
#> $gaps_col
#> [1] 5 10 15